- Myc
-
Myc (c-Myc) is a regulator gene that codes for a transcription factor. In the human genome, Myc is located on chromosome 8 and is believed to regulate expression of 15% of all genes [1] through binding on Enhancer Box sequences (E-boxes) and recruiting histone acetyltransferases (HATs). This means that in addition to its role as a classical transcription factor, Myc also functions to regulate global chromatin structure by regulating histone acetylation both in gene-rich regions and at sites far from any known gene.[2]
A mutated version of Myc is found in many cancers, which causes Myc to be constitutively (persistently) expressed. This leads to the unregulated expression of many genes, some of which are involved in cell proliferation and results in the formation of cancer. A common translocation involving Myc is t(8;14) which is critical to the development of most cases of Burkitt's Lymphoma. A recent study demonstrated that temporary inhibition of Myc selectively kills mouse lung cancer cells, making it a potential cancer drug target.[3]
Contents
Discovery
Myc gene was first discovered in Burkitt's lymphoma patients. In Burkitt's lymphoma, cancer cells show chromosomal translocations, in which Chromosome 8 is frequently involved. Cloning the break-point of the fusion chromosomes revealed a gene that was similar to myelocytomatosis viral oncogene (v-Myc). Thus, the newfound cellular gene was named c-Myc.
Structure
Myc protein belongs to Myc family of transcription factors, which also includes N-Myc and L-Myc genes. Myc family of transcription factors contain bHLH/LZ (basic Helix-Loop-Helix Leucine Zipper) domain. Myc protein, through its bHLH domain can bind to DNA, while the leucine zipper domain allows the dimerization with its partner Max, another bHLH transcription factor.
Myc mRNA contains an IRES (internal ribosome entry site) that allows the RNA to be translated into protein when 5' cap-dependent translation is inhibited, such as during viral infection.
Molecular Function
Myc protein is a transcription factor that activates expression of a great number of genes through binding on consensus sequences (Enhancer Box sequences (E-boxes)) and recruiting histone acetyltransferases (HATs). It can also act as a transcriptional repressor. By binding Miz-1 transcription factor and displacing the p300 co-activator, it inhibits expression of Miz-1 target genes. In addition, myc has a direct role in the control of DNA replication.[4]
Myc is activated upon various mitogenic signals such as Wnt, Shh and EGF (via the MAPK/ERK pathway). By modifying the expression of its target genes, Myc activation results in numerous biological effects. The first to be discovered was its capability to drive cell proliferation (upregulates cyclins, downregulates p21), but it also plays a very important role in regulating cell growth (upregulates ribosomal RNA and proteins), apoptosis (downregulates Bcl-2), differentiation and stem cell self-renewal. Myc is a very strong proto-oncogene and it is very often found to be upregulated in many types of cancers. Myc overexpression stimulates gene amplification,[5] presumably through DNA over-replication.
Animal Models
During the discovery of Myc gene, it was realized that chromosomes that translocate to Chromosome 8 contained immunoglobulin genes at the break-point. Enhancers that normally drive expression of immunoglobin genes now lead to overexpression of Myc proto-oncogene in lymphoma cells. To study the mechanism of tumorigenesis in Burkitt's lymphoma by mimicking expression pattern of Myc in these cancer cells, transgenic mouse models were developed. Myc gene placed under the control of IgM heavy chain enhancer in transgenic mice gives rise to mainly lymphomas. Later on, in order to study effects of Myc in other types of cancer, transgenic mice that overexpress Myc in different tissues (liver, breast) were also made. In all these mouse models overexpression of Myc causes tumorigenesis, illustrating the potency of Myc oncogene.
Interactions
Myc has been shown to interact with NMI,[6] NFYC,[7] NFYB,[8] Cyclin T1,[9] RuvB-like 1,[10][11] GTF2I,[12] BRCA1,[6][13][14][15] T-cell lymphoma invasion and metastasis-inducing protein 1,[16] ACTL6A,[11] PCAF,[17] MYCBP2,[18] MAPK8,[19] Bcl-2,[20] Transcription initiation protein SPT3 homolog,[17] SAP130,[17] DNMT3A,[21] Mothers against decapentaplegic homolog 3,[22] MAX,[23][24][25][26][27][28][29][30][31][32][33][34][35] Mothers against decapentaplegic homolog 2,[22] MYCBP,[36] HTATIP,[37] ZBTB17,[38][39] Transformation/transcription domain-associated protein,[11][17][24][25] TADA2L,[17] PFDN5,[40][41] MAPK1,[20][42][43] TFAP2A,[44] P73,[45] TAF9,[17] YY1,[46] SMARCB1,[26] SMARCA4,[11][23] MLH1[27], EP400[10] and let-7.[47][48][49]
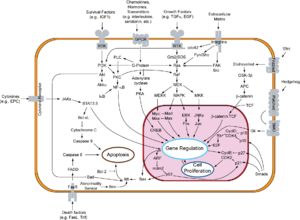 Overview of signal transduction pathways involved in apoptosis.
Overview of signal transduction pathways involved in apoptosis.
Effects
A major effect of Myc is B cell proliferation.[50]
c-Myc induces AEG-1 or MTDH gene expression and in turn itself requires AEG-1 oncogene for its expression.
See also
References
- ^ Gearhart J, Pashos EE, Prasad MK, Pluripotency Redeux -- advances in stem-cell research, N Engl J Med 357(15):1469 Free full text
- ^ Cotterman R, Jin VX, Krig SR, Lemen JM, Wey A, Farnham PJ, Knoepfler PS. (2008). "N-Myc regulates a widespread euchromatic program in the human genome partially independent of its role as a classical transcription factor". Cancer Res. 68 (23): 9654–62. doi:10.1158/0008-5472.CAN-08-1961. PMC 2637654. PMID 19047142. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2637654.
- ^ Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI (October 2008). "Modelling Myc inhibition as a cancer therapy". Nature 455 (7213): 679–83. doi:10.1038/nature07260. PMID 18716624.
- ^ Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R (July 2007). "Non-transcriptional control of DNA replication by c-Myc". Nature 448 (7152): 445–51. doi:10.1038/nature05953. PMID 17597761.
- ^ Denis N, Kitzis A, Kruh J, Dautry F, Corcos D (August 1991). "Stimulation of methotrexate resistance and dihydrofolate reductase gene amplification by c-myc". Oncogene 6 (8): 1453–7. PMID 1886715.
- ^ a b Li, Huchun; Lee Tae-Hee, Avraham Hava (Jun. 2002). "A novel tricomplex of BRCA1, Nmi, and c-Myc inhibits c-Myc-induced human telomerase reverse transcriptase gene (hTERT) promoter activity in breast cancer". J. Biol. Chem. (United States) 277 (23): 20965–73. doi:10.1074/jbc.M112231200. ISSN 0021-9258. PMID 11916966.
- ^ Taira, T; Sawai M, Ikeda M, Tamai K, Iguchi-Ariga S M, Ariga H (Aug. 1999). "Cell cycle-dependent switch of up-and down-regulation of human hsp70 gene expression by interaction between c-Myc and CBF/NF-Y". J. Biol. Chem. (UNITED STATES) 274 (34): 24270–9. doi:10.1074/jbc.274.34.24270. ISSN 0021-9258. PMID 10446203.
- ^ Izumi, H; Molander C, Penn L Z, Ishisaki A, Kohno K, Funa K (Apr. 2001). "Mechanism for the transcriptional repression by c-Myc on PDGF beta-receptor". J. Cell. Sci. (England) 114 (Pt 8): 1533–44. ISSN 0021-9533. PMID 11282029.
- ^ Kanazawa, Satoshi; Soucek Laura, Evan Gerard, Okamoto Takashi, Peterlin B Matija (Aug. 2003). "c-Myc recruits P-TEFb for transcription, cellular proliferation and apoptosis". Oncogene (England) 22 (36): 5707–11. doi:10.1038/sj.onc.1206800. ISSN 0950-9232. PMID 12944920.
- ^ a b Fuchs, M; Gerber J, Drapkin R, Sif S, Ikura T, Ogryzko V, Lane W S, Nakatani Y, Livingston D M (Aug. 2001). "The p400 complex is an essential E1A transformation target". Cell (United States) 106 (3): 297–307. doi:10.1016/S0092-8674(01)00450-0. ISSN 0092-8674. PMID 11509179.
- ^ a b c d Park, Jeonghyeon; Wood Marcelo A, Cole Michael D (Mar. 2002). "BAF53 Forms Distinct Nuclear Complexes and Functions as a Critical c-Myc-Interacting Nuclear Cofactor for Oncogenic Transformation". Mol. Cell. Biol. (United States) 22 (5): 1307–16. doi:10.1128/MCB.22.5.1307-1316.2002. ISSN 0270-7306. PMC 134713. PMID 11839798. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=134713.
- ^ Roy, A L; Carruthers C, Gutjahr T, Roeder R G (Sep. 1993). "Direct role for Myc in transcription initiation mediated by interactions with TFII-I". Nature (ENGLAND) 365 (6444): 359–61. doi:10.1038/365359a0. ISSN 0028-0836. PMID 8377829.
- ^ Xiong, Jingbo; Fan Saijun, Meng Qinghui, Schramm Laura, Wang Chenguang, Bouzahza Boumedienne, Zhou Jinnian, Zafonte Brian, Goldberg Itzhak D, Haddad Bassem R, Pestell Richard G, Rosen Eliot M (Dec. 2003). "BRCA1 Inhibition of Telomerase Activity in Cultured Cells". Mol. Cell. Biol. (United States) 23 (23): 8668–90. doi:10.1128/MCB.23.23.8668-8690.2003. ISSN 0270-7306. PMC 262673. PMID 14612409. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=262673.
- ^ Zhou, Chenyi; Liu Jinsong (Mar. 2003). "Inhibition of human telomerase reverse transcriptase gene expression by BRCA1 in human ovarian cancer cells". Biochem. Biophys. Res. Commun. (United States) 303 (1): 130–6. doi:10.1016/S0006-291X(03)00318-8. ISSN 0006-291X. PMID 12646176.
- ^ Wang, Q; Zhang H, Kajino K, Greene M I (Oct. 1998). "BRCA1 binds c-Myc and inhibits its transcriptional and transforming activity in cells". Oncogene (ENGLAND) 17 (15): 1939–48. doi:10.1038/sj.onc.1202403. ISSN 0950-9232. PMID 9788437.
- ^ Otsuki, Yoshiro; Tanaka Masamitsu, Kamo Takaharu, Kitanaka Chifumi, Kuchino Yoshiyuki, Sugimura Haruhiko (Feb. 2003). "Guanine nucleotide exchange factor, Tiam1, directly binds to c-Myc and interferes with c-Myc-mediated apoptosis in rat-1 fibroblasts". J. Biol. Chem. (United States) 278 (7): 5132–40. doi:10.1074/jbc.M206733200. ISSN 0021-9258. PMID 12446731.
- ^ a b c d e f Liu, Xiaohui; Tesfai Jerusalem, Evrard Yvonne A, Dent Sharon Y R, Martinez Ernest (May. 2003). "c-Myc transformation domain recruits the human STAGA complex and requires TRRAP and GCN5 acetylase activity for transcription activation". J. Biol. Chem. (United States) 278 (22): 20405–12. doi:10.1074/jbc.M211795200. ISSN 0021-9258. PMID 12660246.
- ^ Guo, Q; Xie J, Dang C V, Liu E T, Bishop J M (Aug. 1998). "Identification of a large Myc-binding protein that contains RCC1-like repeats". Proc. Natl. Acad. Sci. U.S.A. (UNITED STATES) 95 (16): 9172–7. doi:10.1073/pnas.95.16.9172. ISSN 0027-8424. PMC 21311. PMID 9689053. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=21311.
- ^ Noguchi, K; Kitanaka C, Yamana H, Kokubu A, Mochizuki T, Kuchino Y (Nov. 1999). "Regulation of c-Myc through phosphorylation at Ser-62 and Ser-71 by c-Jun N-terminal kinase". J. Biol. Chem. (UNITED STATES) 274 (46): 32580–7. doi:10.1074/jbc.274.46.32580. ISSN 0021-9258. PMID 10551811.
- ^ a b Jin, Zhaohui; Gao Fengqin, Flagg Tammy, Deng Xingming (Sep. 2004). "Tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone promotes functional cooperation of Bcl2 and c-Myc through phosphorylation in regulating cell survival and proliferation". J. Biol. Chem. (United States) 279 (38): 40209–19. doi:10.1074/jbc.M404056200. ISSN 0021-9258. PMID 15210690.
- ^ Brenner, Carmen; Deplus Rachel, Didelot Céline, Loriot Axelle, Viré Emmanuelle, De Smet Charles, Gutierrez Arantxa, Danovi Davide, Bernard David, Boon Thierry, Pelicci Pier Giuseppe, Amati Bruno, Kouzarides Tony, de Launoit Yvan, Di Croce Luciano, Fuks François (Jan. 2005). "Myc represses transcription through recruitment of DNA methyltransferase corepressor". EMBO J. (England) 24 (2): 336–46. doi:10.1038/sj.emboj.7600509. ISSN 0261-4189. PMC 545804. PMID 15616584. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=545804.
- ^ a b Feng, Xin-Hua; Liang Yao-Yun, Liang Min, Zhai Weiguo, Lin Xia (Jan. 2002). "Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-beta-mediated induction of the CDK inhibitor p15(Ink4B)". Mol. Cell (United States) 9 (1): 133–43. doi:10.1016/S1097-2765(01)00430-0. ISSN 1097-2765. PMID 11804592.
- ^ a b Ewing, Rob M; Chu Peter, Elisma Fred, Li Hongyan, Taylor Paul, Climie Shane, McBroom-Cerajewski Linda, Robinson Mark D, O'Connor Liam, Li Michael, Taylor Rod, Dharsee Moyez, Ho Yuen, Heilbut Adrian, Moore Lynda, Zhang Shudong, Ornatsky Olga, Bukhman Yury V, Ethier Martin, Sheng Yinglun, Vasilescu Julian, Abu-Farha Mohamed, Lambert Jean-Philippe, Duewel Henry S, Stewart Ian I, Kuehl Bonnie, Hogue Kelly, Colwill Karen, Gladwish Katharine, Muskat Brenda, Kinach Robert, Adams Sally-Lin, Moran Michael F, Morin Gregg B, Topaloglou Thodoros, Figeys Daniel (2007). "Large-scale mapping of human protein–protein interactions by mass spectrometry". Mol. Syst. Biol. (England) 3 (1): 89. doi:10.1038/msb4100134. PMC 1847948. PMID 17353931. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1847948.
- ^ a b McMahon, S B; Wood M A, Cole M D (Jan. 2000). "The Essential Cofactor TRRAP Recruits the Histone Acetyltransferase hGCN5 to c-Myc". Mol. Cell. Biol. (UNITED STATES) 20 (2): 556–62. doi:10.1128/MCB.20.2.556-562.2000. ISSN 0270-7306. PMC 85131. PMID 10611234. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=85131.
- ^ a b McMahon, S B; Van Buskirk H A, Dugan K A, Copeland T D, Cole M D (Aug. 1998). "The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins". Cell (UNITED STATES) 94 (3): 363–74. doi:10.1016/S0092-8674(00)81479-8. ISSN 0092-8674. PMID 9708738.
- ^ a b Cheng, S W; Davies K P, Yung E, Beltran R J, Yu J, Kalpana G V (May. 1999). "c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function". Nat. Genet. (UNITED STATES) 22 (1): 102–5. doi:10.1038/8811. ISSN 1061-4036. PMID 10319872.
- ^ a b Mac Partlin, Mary; Homer Elizabeth, Robinson Helen, McCormick Carol J, Crouch Dorothy H, Durant Stephen T, Matheson Elizabeth C, Hall Andrew G, Gillespie David A F, Brown Robert (Feb. 2003). "Interactions of the DNA mismatch repair proteins MLH1 and MSH2 with c-MYC and MAX". Oncogene (England) 22 (6): 819–25. doi:10.1038/sj.onc.1206252. ISSN 0950-9232. PMID 12584560.
- ^ Blackwood, E M; Eisenman R N (Mar. 1991). "Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc". Science (UNITED STATES) 251 (4998): 1211–7. doi:10.1126/science.2006410. ISSN 0036-8075. PMID 2006410.
- ^ Lee, Clement M; Onésime Djamila, Reddy C Damodara, Dhanasekaran N, Reddy E Premkumar (Oct. 2002). "JLP: A scaffolding protein that tethers JNK/p38MAPK signaling modules and transcription factors". Proc. Natl. Acad. Sci. U.S.A. (United States) 99 (22): 14189–94. doi:10.1073/pnas.232310199. ISSN 0027-8424. PMC 137859. PMID 12391307. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=137859.
- ^ Billin, A N; Eilers A L, Queva C, Ayer D E (Dec. 1999). "Mlx, a novel Max-like BHLHZip protein that interacts with the Max network of transcription factors". J. Biol. Chem. (UNITED STATES) 274 (51): 36344–50. doi:10.1074/jbc.274.51.36344. ISSN 0021-9258. PMID 10593926.
- ^ Gupta, K; Anand G, Yin X, Grove L, Prochownik E V (Mar. 1998). "Mmip1: a novel leucine zipper protein that reverses the suppressive effects of Mad family members on c-myc". Oncogene (ENGLAND) 16 (9): 1149–59. doi:10.1038/sj.onc.1201634. ISSN 0950-9232. PMID 9528857.
- ^ Meroni, G; Reymond A, Alcalay M, Borsani G, Tanigami A, Tonlorenzi R, Lo Nigro C, Messali S, Zollo M, Ledbetter D H, Brent R, Ballabio A, Carrozzo R (May. 1997). "Rox, a novel bHLHZip protein expressed in quiescent cells that heterodimerizes with Max, binds a non-canonical E box and acts as a transcriptional repressor". EMBO J. (ENGLAND) 16 (10): 2892–906. doi:10.1093/emboj/16.10.2892. ISSN 0261-4189. PMC 1169897. PMID 9184233. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1169897.
- ^ Nair, Satish K; Burley Stephen K (Jan. 2003). "X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors". Cell (United States) 112 (2): 193–205. doi:10.1016/S0092-8674(02)01284-9. ISSN 0092-8674. PMID 12553908.
- ^ FitzGerald, M J; Arsura M, Bellas R E, Yang W, Wu M, Chin L, Mann K K, DePinho R A, Sonenshein G E (Apr. 1999). "Differential effects of the widely expressed dMax splice variant of Max on E-box vs initiator element-mediated regulation by c-Myc". Oncogene (ENGLAND) 18 (15): 2489–98. doi:10.1038/sj.onc.1202611. ISSN 0950-9232. PMID 10229200.
- ^ Meroni, G; Cairo S, Merla G, Messali S, Brent R, Ballabio A, Reymond A (Jul. 2000). "Mlx, a new Max-like bHLHZip family member: the center stage of a novel transcription factors regulatory pathway?". Oncogene (ENGLAND) 19 (29): 3266–77. doi:10.1038/sj.onc.1203634. ISSN 0950-9232. PMID 10918583.
- ^ Taira, T; Maëda J, Onishi T, Kitaura H, Yoshida S, Kato H, Ikeda M, Tamai K, Iguchi-Ariga S M, Ariga H (Aug. 1998). "AMY-1, a novel C-MYC binding protein that stimulates transcription activity of C-MYC". Genes Cells (ENGLAND) 3 (8): 549–65. doi:10.1046/j.1365-2443.1998.00206.x. ISSN 1356-9597. PMID 9797456.
- ^ Frank, Scott R; Parisi Tiziana, Taubert Stefan, Fernandez Paula, Fuchs Miriam, Chan Ho-Man, Livingston David M, Amati Bruno (Jun. 2003). "MYC recruits the TIP60 histone acetyltransferase complex to chromatin". EMBO Rep. (England) 4 (6): 575–80. doi:10.1038/sj.embor.embor861. ISSN 1469-221X. PMC 1319201. PMID 12776177. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1319201.
- ^ Staller, P; Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, Möröy T, Bartek J, Massagué J, Hänel F, Eilers M (Apr. 2001). "Repression of p15INK4b expression by Myc through association with Miz-1". Nat. Cell Biol. (England) 3 (4): 392–9. doi:10.1038/35070076. ISSN 1465-7392. PMID 11283613.
- ^ Peukert, K; Staller P, Schneider A, Carmichael G, Hänel F, Eilers M (Sep. 1997). "An alternative pathway for gene regulation by Myc". EMBO J. (ENGLAND) 16 (18): 5672–86. doi:10.1093/emboj/16.18.5672. ISSN 0261-4189. PMC 1170199. PMID 9312026. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1170199.
- ^ Mori, K; Maeda Y, Kitaura H, Taira T, Iguchi-Ariga S M, Ariga H (Nov. 1998). "MM-1, a novel c-Myc-associating protein that represses transcriptional activity of c-Myc". J. Biol. Chem. (UNITED STATES) 273 (45): 29794–800. doi:10.1074/jbc.273.45.29794. ISSN 0021-9258. PMID 9792694.
- ^ Fujioka, Y; Taira T, Maeda Y, Tanaka S, Nishihara H, Iguchi-Ariga S M, Nagashima K, Ariga H (Nov. 2001). "MM-1, a c-Myc-binding protein, is a candidate for a tumor suppressor in leukemia/lymphoma and tongue cancer". J. Biol. Chem. (United States) 276 (48): 45137–44. doi:10.1074/jbc.M106127200. ISSN 0021-9258. PMID 11567024.
- ^ Gupta, S; Davis R J (Oct. 1994). "MAP kinase binds to the NH2-terminal activation domain of c-Myc". FEBS Lett. (NETHERLANDS) 353 (3): 281–5. doi:10.1016/0014-5793(94)01052-8. ISSN 0014-5793. PMID 7957875.
- ^ Tournier, C; Whitmarsh A J, Cavanagh J, Barrett T, Davis R J (Jul. 1997). "Mitogen-activated protein kinase kinase 7 is an activator of the c-Jun NH2-terminal kinase". Proc. Natl. Acad. Sci. U.S.A. (UNITED STATES) 94 (14): 7337–42. doi:10.1073/pnas.94.14.7337. ISSN 0027-8424. PMC 23822. PMID 9207092. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=23822.
- ^ Gaubatz, S; Imhof A, Dosch R, Werner O, Mitchell P, Buettner R, Eilers M (Apr. 1995). "Transcriptional activation by Myc is under negative control by the transcription factor AP-2". EMBO J. (ENGLAND) 14 (7): 1508–19. ISSN 0261-4189. PMC 398238. PMID 7729426. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=398238.
- ^ Uramoto, Hidetaka; Izumi Hiroto, Ise Tomoko, Tada Mitsuhiro, Uchiumi Takeshi, Kuwano Michihiko, Yasumoto Kosei, Funa Keiko, Kohno Kimitoshi (Aug. 2002). "p73 Interacts with c-Myc to regulate Y-box-binding protein-1 expression". J. Biol. Chem. (United States) 277 (35): 31694–702. doi:10.1074/jbc.M200266200. ISSN 0021-9258. PMID 12080043.
- ^ Shrivastava, A; Saleque S, Kalpana G V, Artandi S, Goff S P, Calame K (Dec. 1993). "Inhibition of transcriptional regulator Yin-Yang-1 by association with c-Myc". Science (UNITED STATES) 262 (5141): 1889–92. doi:10.1126/science.8266081. ISSN 0036-8075. PMID 8266081.
- ^ Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT (January 2008). "Widespread microRNA repression by Myc contributes to tumorigenesis". Nat. Genet. 40 (1): 43–50. doi:10.1038/ng.2007.30. PMC 2628762. PMID 18066065. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2628762.
- ^ Koscianska E, Baev V, Skreka K, Oikonomaki K, Rusinov V, Tabler M, Kalantidis K (2007). "Prediction and preliminary validation of oncogene regulation by miRNAs". BMC Mol. Biol. 8: 79. doi:10.1186/1471-2199-8-79. PMC 2096627. PMID 17877811. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2096627.
- ^ Ioannidis P, Mahaira LG, Perez SA, Gritzapis AD, Sotiropoulou PA, Kavalakis GJ, Antsaklis AI, Baxevanis CN, Papamichail M (May 2005). "CRD-BP/IMP1 expression characterizes cord blood CD34+ stem cells and affects c-myc and IGF-II expression in MCF-7 cancer cells". J. Biol. Chem. 280 (20): 20086–93. doi:10.1074/jbc.M410036200. PMID 15769738.
- ^ de Alboran IM, O'Hagan RC, Gärtner F, et al. (January 2001). "Analysis of C-MYC function in normal cells via conditional gene-targeted mutation". Immunity 14 (1): 45–55. doi:10.1016/S1074-7613(01)00088-7. PMID 11163229.
Further reading
- Ruf IK, Rhyne PW, Yang H, et al. (2002). "EBV regulates c-MYC, apoptosis, and tumorigenicity in Burkitt's lymphoma". Curr. Top. Microbiol. Immunol. 258: 153–60. PMID 11443860.
- Lüscher B (2001). "Function and regulation of the transcription factors of the Myc/Max/Mad network". Gene 277 (1–2): 1–14. doi:10.1016/S0378-1119(01)00697-7. PMID 11602341.
- Hoffman B, Amanullah A, Shafarenko M, Liebermann DA (2002). "The proto-oncogene c-myc in hematopoietic development and leukemogenesis". Oncogene 21 (21): 3414–21. doi:10.1038/sj.onc.1205400. PMID 12032779.
- Pelengaris S, Khan M, Evan G (2002). "c-MYC: more than just a matter of life and death". Nat. Rev. Cancer 2 (10): 764–76. doi:10.1038/nrc904. PMID 12360279.
- Nilsson JA, Cleveland JL (2004). "Myc pathways provoking cell suicide and cancer". Oncogene 22 (56): 9007–21. doi:10.1038/sj.onc.1207261. PMID 14663479.
- Dang CV, O'donnell KA, Juopperi T (2005). "The great MYC escape in tumorigenesis". Cancer Cell 8 (3): 177–8. doi:10.1016/j.ccr.2005.08.005. PMID 16169462.
- Dang CV, Li F, Lee LA (2007). "Could MYC induction of mitochondrial biogenesis be linked to ROS production and genomic instability?". Cell Cycle 4 (11): 1465–6. doi:10.4161/cc.4.11.2121. PMID 16205115.
- Coller HA, Forman JJ, Legesse-Miller A (2007). ""Myc'ed Messages": Myc Induces Transcription of E2F1 while Inhibiting Its Translation via a microRNA Polycistron". PLoS Genet. 3 (8): e146. doi:10.1371/journal.pgen.0030146. PMC 1959363. PMID 17784791. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1959363.
- Astrin SM, Laurence J (1992). "Human immunodeficiency virus activates c-myc and Epstein-Barr virus in human B lymphocytes". Ann. N. Y. Acad. Sci. 651: 422–32. doi:10.1111/j.1749-6632.1992.tb24642.x. PMID 1318011.
- Bernstein PL, Herrick DJ, Prokipcak RD, Ross J (1992). "Control of c-myc mRNA half-life in vitro by a protein capable of binding to a coding region stability determinant". Genes Dev. 6 (4): 642–54. doi:10.1101/gad.6.4.642. PMID 1559612.
- Iijima S, Teraoka H, Date T, Tsukada K (1992). "DNA-activated protein kinase in Raji Burkitt's lymphoma cells. Phosphorylation of c-Myc oncoprotein". Eur. J. Biochem. 206 (2): 595–603. doi:10.1111/j.1432-1033.1992.tb16964.x. PMID 1597196.
- Seth A, Alvarez E, Gupta S, Davis RJ (1992). "A phosphorylation site located in the NH2-terminal domain of c-Myc increases transactivation of gene expression". J. Biol. Chem. 266 (35): 23521–4. PMID 1748630.
- Takahashi E, Hori T, O'Connell P, et al. (1991). "Mapping of the MYC gene to band 8q24.12----q24.13 by R-banding and distal to fra(8)(q24.11), FRA8E, by fluorescence in situ hybridization". Cytogenet. Cell Genet. 57 (2–3): 109–11. doi:10.1159/000133124. PMID 1914517.
- Blackwood EM, Eisenman RN (1991). "Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc". Science 251 (4998): 1211–7. doi:10.1126/science.2006410. PMID 2006410.
- Gazin C, Rigolet M, Briand JP, et al. (1986). "Immunochemical detection of proteins related to the human c-myc exon 1". EMBO J. 5 (9): 2241–50. PMC 1167107. PMID 2430795. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1167107.
- Lüscher B, Kuenzel EA, Krebs EG, Eisenman RN (1989). "Myc oncoproteins are phosphorylated by casein kinase II". EMBO J. 8 (4): 1111–9. PMC 400922. PMID 2663470. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=400922.
- Finver SN, Nishikura K, Finger LR, et al. (1988). "Sequence analysis of the MYC oncogene involved in the t(8;14)(q24;q11) chromosome translocation in a human leukemia T-cell line indicates that putative regulatory regions are not altered". Proc. Natl. Acad. Sci. U.S.A. 85 (9): 3052–6. doi:10.1073/pnas.85.9.3052. PMC 280141. PMID 2834731. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=280141.
- Showe LC, Moore RC, Erikson J, Croce CM (1987). "MYC oncogene involved in a t(8;22) chromosome translocation is not altered in its putative regulatory regions". Proc. Natl. Acad. Sci. U.S.A. 84 (9): 2824–8. doi:10.1073/pnas.84.9.2824. PMC 304752. PMID 3033665. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=304752.
- Guilhot S, Petridou B, Syed-Hussain S, Galibert F (1989). "Nucleotide sequence 3' to the human c-myc oncogene; presence of a long inverted repeat". Gene 72 (1–2): 105–8. doi:10.1016/0378-1119(88)90131-X. PMID 3243428.
- Hann SR, King MW, Bentley DL, et al. (1988). "A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt's lymphomas". Cell 52 (2): 185–95. doi:10.1016/0092-8674(88)90507-7. PMID 3277717.
External links
- The Myc Protein
- NCBI Human Myc protein
- Myc cancer gene
- MeSH myc+Proto-Oncogene+Proteins
- Generating iPS Cells from MEFS through Forced Expression of Sox-2, Oct-4, c-Myc, and Klf4
- Drosophila Myc - The Interactive Fly
- FactorBook C-Myc
PDB gallery Neoplasm: Tumor suppressor genes/proteins and Oncogenes/Proto-oncogenes Ligand Receptor TSP: CDH1TSP: PTCH1TSP: TGF beta receptor 2Intracellular signaling P+Ps ONCO: Beta-catenin · TSP: APCHippo signaling pathwayOther/unknownNucleus TSP: VHL · ONCO: CBL - MDM2Mitochondria Other/ungrouped M: NEO
tsoc, mrkr
tumr, epon, para
drug (L1i/1e/V03)
Transcription factors and intracellular receptors (1) Basic domains (1.1) Basic leucine zipper (bZIP)Activating transcription factor (AATF, 1, 2, 3, 4, 5, 6, 7) · AP-1 (c-Fos, FOSB, FOSL1, FOSL2, JDP2, c-Jun, JUNB, JUND) · BACH (1, 2) · BATF · BLZF1 · C/EBP (α, β, γ, δ, ε, ζ) · CREB (1, 3, L1) · CREM · DBP · DDIT3 · GABPA · HLF · MAF (B, F, G, K) · NFE (2, L1, L2, L3) · NFIL3 · NRL · NRF (1, 2, 3) · XBP1(1.2) Basic helix-loop-helix (bHLH)ATOH1 · AhR · AHRR · ARNT · ASCL1 · BHLHB2 · BMAL (ARNTL, ARNTL2) · CLOCK · EPAS1 · FIGLA · HAND (1, 2) · HES (5, 6) · HEY (1, 2, L) · HES1 · HIF (1A, 3A) · ID (1, 2, 3, 4) · LYL1 · MESP2 · MXD4 · MYCL1 · MYCN · Myogenic regulatory factors (MyoD, Myogenin, MYF5, MYF6) · Neurogenins (1, 2, 3) · NeuroD (1, 2) · NPAS (1, 2, 3) · OLIG (1, 2) · Pho4 · Scleraxis · SIM (1, 2) · TAL (1, 2) · Twist · USF1(1.3) bHLH-ZIP(1.4) NF-1(1.5) RF-X(1.6) Basic helix-span-helix (bHSH)(2) Zinc finger DNA-binding domains (2.1) Nuclear receptor (Cys4)subfamily 1 (Thyroid hormone (α, β), CAR, FXR, LXR (α, β), PPAR (α, β/δ, γ), PXR, RAR (α, β, γ), ROR (α, β, γ), Rev-ErbA (α, β), VDR)
subfamily 2 (COUP-TF (I, II), Ear-2, HNF4 (α, γ), PNR, RXR (α, β, γ), Testicular receptor (2, 4), TLX)
subfamily 3 (Steroid hormone (Androgen, Estrogen (α, β), Glucocorticoid, Mineralocorticoid, Progesterone), Estrogen related (α, β, γ))
subfamily 4 NUR (NGFIB, NOR1, NURR1) · subfamily 5 (LRH-1, SF1) · subfamily 6 (GCNF) · subfamily 0 (DAX1, SHP)(2.2) Other Cys4(2.3) Cys2His2General transcription factors (TFIIA, TFIIB, TFIID, TFIIE (1, 2), TFIIF (1, 2), TFIIH (1, 2, 4, 2I, 3A, 3C1, 3C2))
ATBF1 · BCL (6, 11A, 11B) · CTCF · E4F1 · EGR (1, 2, 3, 4) · ERV3 · GFI1 · GLI-Krüppel family (1, 2, 3, REST, S2, YY1) · HIC (1, 2) · HIVEP (1, 2, 3) · IKZF (1, 2, 3) · ILF (2, 3) · KLF (2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 17) · MTF1 · MYT1 · OSR1 · PRDM9 · SALL (1, 2, 3, 4) · SP (1, 2, 4, 7, 8) · TSHZ3 · WT1 · Zbtb7 (7A, 7B) · ZBTB (16, 17, 20, 32, 33, 40) · zinc finger (3, 7, 9, 10, 19, 22, 24, 33B, 34, 35, 41, 43, 44, 51, 74, 143, 146, 148, 165, 202, 217, 219, 238, 239, 259, 267, 268, 281, 295, 300, 318, 330, 346, 350, 365, 366, 384, 423, 451, 452, 471, 593, 638, 644, 649, 655)(2.4) Cys6(2.5) Alternating composition(3) Helix-turn-helix domains (3.1) HomeodomainARX · CDX (1, 2) · CRX · CUTL1 · DBX (1, 2) · DLX (3, 4, 5) · EMX2 · EN (1, 2) · FHL (1, 2, 3) · HESX1 · HHEX · HLX · Homeobox (A1, A2, A3, A4, A5, A7, A9, A10, A11, A13, B1, B2, B3, B4, B5, B6, B7, B8, B9, B13, C4, C5, C6, C8, C9, C10, C11, C12, C13, D1, D3, D4, D8, D9, D10, D11, D12, D13) · HOPX · IRX (1, 2, 3, 4, 5, 6, MKX) · LMX (1A, 1B) · MEIS (1, 2) · MEOX2 · MNX1 · MSX (1, 2) · NANOG · NKX (2-1, 2-2, 2-3, 2-5, 3-1, 3-2, 6-1, 6-2) · NOBOX · PBX (1, 2, 3) · PHF (1, 3, 6, 8, 10, 16, 17, 20, 21A) · PHOX (2A, 2B) · PITX (1, 2, 3) · POU domain (PIT-1, BRN-3: A, B, C, Octamer transcription factor: 1, 2, 3/4, 6, 7, 11) · OTX (1, 2) · PDX1 · SATB2 · SHOX2 · VAX1 · ZEB (1, 2)(3.2) Paired box(3.3) Fork head / winged helix(3.4) Heat Shock Factors(3.5) Tryptophan clusters(3.6) TEA domain(4) β-Scaffold factors with minor groove contacts (4.1) Rel homology region(4.2) STAT(4.3) p53(4.4) MADS box(4.6) TATA binding proteins(4.7) High-mobility group(4.10) Cold-shock domainCSDA, YBX1(4.11) Runt(0) Other transcription factors (0.2) HMGI(Y)(0.3) Pocket domain(0.6) MiscellaneousCategories:- Human proteins
- Oncogenes
- Transcription factors
Wikimedia Foundation. 2010.


