- Centrosome
-
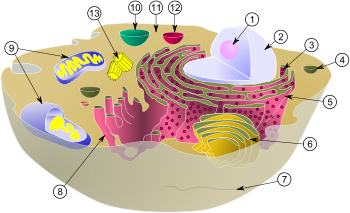 Schematic of typical animal cell, showing subcellular components. Organelles:
Schematic of typical animal cell, showing subcellular components. Organelles:
(1) Nucleolus
(2) Nucleus
(3) Ribosomes (little dots)
(4) Vesicle
(5) Rough endoplasmic reticulum (ER)
(6) Golgi apparatus
(7) Cytoskeleton
(8) Smooth ER
(9) Mitochondria
(10) Vacuole
(11) Cytoplasm
(12) Lysosome
(13) Centrioles within CentrosomeIn cell biology, the centrosome is an organelle that serves as the main microtubule organizing center (MTOC) of the animal cell as well as a regulator of cell-cycle progression. It was discovered by Edouard Van Beneden in 1883 [1] and was described and named in 1888 by Theodor Boveri.[2] In the theory of evolution the centrosome is thought to have evolved only in the metazoan lineage of eukaryotic cells.[3] Fungi and plants use other MTOC structures to organize their microtubules.[4][5] Although the centrosome has a key role in efficient mitosis in animal cells, it is not essential.[6]
Centrosomes are composed of two orthogonally arranged centrioles surrounded by an amorphous mass of protein termed the pericentriolar material (PCM). The PCM contains proteins responsible for microtubule nucleation and anchoring[7] including γ-tubulin, pericentrin and ninein. In general, each centriole of the centrosome is based on a nine triplet microtubule assembled in a cartwheel structure, and contains centrin, cenexin and tektin.[8]
Contents
Roles of the centrosome
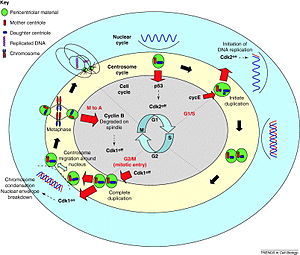
Centrosomes are associated with the nuclear membrane during prophase of the cell cycle. In mitosis the nuclear membrane breaks down and the centrosome nucleated microtubules can interact with the chromosomes to build the mitotic spindle.
The mother centriole, the one that was inherited from the mother cell, also has a central role in making cilia and flagella.[8]
The centrosome is copied only once per cell cycle so that each daughter cell inherits one centrosome, containing two centrioles (see also: centrosome cycle). The centrosome replicates during the S phase of the cell cycle. During the prophase in the process of cell division called mitosis, the centrosomes migrate to opposite poles of the cell. The mitotic spindle then forms between the two centrosomes. Upon division, each daughter cell receives one centrosome. Aberrant numbers of centrosomes in a cell have been associated with cancer. Doubling of a centrosome is similar to DNA replication in two respects: the semiconservative nature of the process and the action of cdk2 as a regulator of the process.[9] But the processes are essentially different in that centrosome doubling does not occur by template reading and assembly. The mother centriole just aids in the accumulation of materials required for the assembly of the daughter centriole.[10]
In animal cells, centrosomes contain two structures called centrioles. Interestingly, centrioles are not required for the progression of mitosis. When the centrioles are irradiated by a laser, mitosis proceeds normally with a morphologically normal spindle. Moreover, development of the fruit fly Drosophila is largely normal when centrioles are absent due to a mutation in a gene required for their duplication.[11] In the absence of the centrioles the microtubules of the spindle are focused by motors allowing the formation of a bipolar spindle. Many cells can completely undergo interphase without centrioles.[8] Unlike centrioles, centrosomes are required for survival of the organism. Acentrosomal cells lack radial arrays of astral microtubules. They are also defective in spindle positioning and in ability to establish a central localization site in cytokinesis. The function of centrosome in this context is hypothesized to ensure the fidelity of cell division because it greatly increases the efficacy. Some cell types arrest in the following cell cycle when centrosomes are absent. This is not a universal phenomenon.
When the nematode C. elegans egg is fertilized the sperm delivers a pair of centrioles. These centrioles will form the centrosomes which will direct the first cell division of the zygote and this will determine its polarity. It is not yet clear whether the role of the centrosome in polarity determination is microtubule dependent or independent.
Centrosome alterations in cancer cells
It was very early identified (at the end of the 19th century) that centrosomes are frequently altered in cancer cells, by Theodor Boveri, the scientist who named the centrosome itself. This initial observation was subsequently extended to many types of human tumors.[12] Centrosome alterations in cancer can be divided in two subgroups, structural or numeric aberrations, yet both can be simultaneously found in a tumor.
Structural aberrations
Usually they appear due to uncontrolled expression of centrosome components, or due to post-translational modifications (such as phosphorylations) which are not adequate for those components. These modifications may produce variations in centrosome size (usually too big, due to an excess of pericentriolar material). On top of this, due to the fact that centrosomal proteins have the tendency to form aggregates, often centrosome-related bodies (CRBs) are observed in ectopic places. [13] Both enlarged centrosomes and CRBs are similar to the centrosomal structures observed in tumors,[14]. Even more, these structures can be induced in culture cells by overexpression of specific centrosomal proteins, such as CNap-1 or Nlp.[13][15]These structures may look very similar, yet detailed studies reveal that they may present very different properties, depending on their proteic composition. For instance, their capacity to incorporate γ-TuRC complexes (see also: γ-tubulin) can be very variable, and so their capacity to nucleate microtubules,[14] therefore affecting in different way the shape, polarity and motility of implicated tumor cells.
Numeric aberrations
The presence of an inadecuate number of centrosomes is very often linked to the apparition of genome instability and the loss of tissular differentiation.[14][16] However, the method to count the centrosome number (each one with 2 centrioles) is often not very precise, because it is frequently assesed using fluorescence microscopy, whose resolution capacity is not the best. Nevertheless, it is clear that the presence of supernumerary (in excess) centrosomes is a common event in human tumors. It has been observed that loss of the tumor-suppressor protein p53 produces supernumerary centrosomes,[17] as well as deregulation of other proteins implicated in tumorigenesis in humans, such as BRCA1 and BRCA2 (for references, see [12]). It is important to note that supernumerary centrosomes can be generated by very different mechanisms: specific reduplication of the centrosome, failure during cell division (generating an increase in chromosome number), cell fusion (for instance due to infection by specific viruses) or de novo generation of centrosomes. At this point there is not sufficient information to know how frequent are those mechanisms in vivo, but it is possible that the increase in centrosome numbers due a failure during cell division might be more frequent than appreciated, because many "primary" defects in one cell (deregulation of the cell cycle, defective DNA or chromatin metabolism, failure in the spindle checkpoint, etc...) would generate a failure in cell division, an increase in ploidy and an increase in centrosome numbers as a "secondary" effect.[18][19]
Evolution of the centrosome
The evolutionary history of the centrosome and the centriole has been traced for some of the signature genes, e.g. the centrins.[3] Centrins participate in calcium signaling and are required for centriole duplication.[20] There exist two main subfamilies of centrins, both of which are present in the early-branching eukaryote Giardia intestinalis. Centrins have therefore been present in the common ancestor of eukaryotes. Conversely, they have no recognizable homologs in archea and bacteria and are thus part of the "eukaryotic signature genes." Although there are studies on the evolution of the centrins and centrioles,[3][21] no studies have been published on the evolution of the pericentriolar material.
It is evident that some parts of the centrosome are highly diverged in the model species Drosophila melanogaster and Caenorhabditis elegans. For example, both species have lost one of the centrin subfamilies that are usually associated with centriole duplication. Drosophila melanogaster mutants that lack centrosomes can even develop to morphologically normal adult flies, which then die shortly after birth because their sensory neurons lack cilia.[11] Thus, these flies have evolved functionally redundant machinery, which is independent of the centrosomes.
Centrosome associated nucleotides
Research in 2006[22] indicated that centrosomes from Surf clam eggs contain RNA sequences. The sequences identified were found in "few to no" other places in the cell, and do not appear in existing genome databases. One identified RNA sequence contains a putative RNA polymerase, leading to the hypothesis of an RNA based genome within the centrosome. However, subsequent research has shown that centrosome do not contain their own DNA-based genomes. While it was confirmed that RNA molecules associate with centrosomes, the sequences have still been found within the nucleus. Furthermore, centrosomes can form de novo after having been removed (e.g. by laser irradiation) from normal cells.[21]
References
- ^ Wunderlich, V. (2002). "JMM - Past and Present". Journal of Molecular Medicine 80 (9): 545–548. doi:10.1007/s00109-002-0374-y. PMID 12226736.
- ^ Boveri, Theodor (1888). Zellen-Studien II: Die Befruchtung und Teilung des Eies von Ascaris megalocephala.. Jena: Gustav Fischer Verlag. http://www.biodiversitylibrary.org/item/29952.
- ^ a b c Bornens, M.; Azimzadeh, J. (2007). Origin and Evolution of the Centrosome. "Eukaryotic Membranes and Cytoskeleton". Advances in experimental medicine and biology. Advances in Experimental Medicine and Biology 607: 119–129. doi:10.1007/978-0-387-74021-8_10. ISBN 978-0-387-74020-1. PMID 17977464.
- ^ Schmit (2002). "Acentrosomal microtubule nucleation in higher plants". International review of cytology. International Review of Cytology 220: 257–289. doi:10.1016/S0074-7696(02)20008-X. ISBN 9780123646248. PMID 12224551.
- ^ Jaspersen, S. L.; Winey, M. (2004). "THE BUDDING YEAST SPINDLE POLE BODY: Structure, Duplication, and Function". Annual Review of Cell and Developmental Biology 20 (1): 1–28. doi:10.1146/annurev.cellbio.20.022003.114106. PMID 15473833.
- ^ Mahoney, N. M.; Goshima, G.; Douglass, A. D.; Vale, R. D. (2006). "Making Microtubules and Mitotic Spindles in Cells without Functional Centrosomes". Current Biology 16 (6): 564–569. doi:10.1016/j.cub.2006.01.053. PMID 16546079.
- ^ Eddé, B; Rossier; Le Caer; Desbruyères; Gros; Denoulet (1990). "Posttranslational glutamylation of alpha-tubulin". Science 247 (4938): 83–5. Bibcode 1990Sci...247...83E. doi:10.1126/science.1967194. PMID 1967194.
- ^ a b c Rieder, C. F. (Oct 2001). "The centrosome in vertebrates: more than a microtubule-organizing center". Trends in cell biology 11 (10): 413–419. doi:10.1016/S0962-8924(01)02085-2. ISSN 0962-8924. PMID 11567874.
- ^ Stearns, T. (May 2001). "Centrosome duplication. A centriolar pas de deux". Cell 105 (4): 417–420. doi:10.1016/S0092-8674(01)00366-X. ISSN 0092-8674. PMID 11371338.
- ^ Rodrigues-martins, A.; Riparbelli, M.; Callaini, G.; Glover, D. M.; Bettencourt-dias, M. (2007). "Revisiting the Role of the Mother Centriole in Centriole Biogenesis". Science 316 (5827): 1046. Bibcode 2007Sci...316.1046R. doi:10.1126/science.1142950. PMID 17463247.
- ^ a b Basto, R.; Lau, J.; Vinogradova, T.; Gardiol, A.; Woods, G.; Khodjakov, A.; Raff, W. (Jun 2006). "Flies without centrioles". Cell 125 (7): 1375–1386. doi:10.1016/j.cell.2006.05.025. ISSN 0092-8674. PMID 16814722.
- ^ a b Nigg, E.A. (2002). "Centrosome aberrations: cause or consequence of cancer progression?". Nat Rev Cancer 2 (11): 815–821. doi:10.1038/nrc924. PMID 12415252
- ^ a b Casenghi, M.; Meraldi, P.; Weinhart, U.; Duncan, P.I.; Korner, R.; Nigg, E.A. (2003). "Polo-like kinase 1 regulates Nlp, a centrosome protein involved in microtubule nucleation". Dev Cell 5 (1): 113–125. doi:10.1016/S1534-5807(03)00193-X. PMID 12852856
- ^ a b c Lingle, W.L.; Barrett, S.L.; Negron, V.C.; D'assoro, A.B.; Boeneman, K.; Liu W. Whitehead C.M.; Reynolds, C.; Salisbury, J.L. et al. (2002). "Centrosome amplification drives chromosomal instability in breast tumor development". Proc Natl Acad Sci USA 99 (4): 1978–1983. doi:10.1073/pnas.032479999. PMC 122305. PMID 11830638. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=122305
- ^ Fry, A.M.; Mayor, T.; Meraldi, P.; Stierhof, Y.D.; Tanaka, K.; Nigg, E.A. (1998). "C-Nap1, a Novel Centrosomal Coiled-Coil Protein and Candidate Substrate of the Cell Cycle–regulated Protein Kinase Nek2". J Cell Biol 141 (7): 1563–1574. doi:10.1083/jcb.141.7.1563. PMC 2133000. PMID 9647649. http://jcb.rupress.org/cgi/content/abstract/141/7/1563. Retrieved 2009-07-29
- ^ Ghadimi, B.M.; Sackett, D.L.; Difilippantonio, M.J.; Schrock, E.; Neumann, T.; Jauho, A.; Auer, G.; Ried, T. (2000). "Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations". Genes Chromosomes Cancer 27 (2): 183–190. doi:10.1002/(SICI)1098-2264(200002)27:2<183::AID-GCC10>3.0.CO;2-P. PMID 10612807
- ^ Fukasawa, K.; Choi, T.; Kuriyama, R.; Rulong, S.; Woude, Vande G.F. (1996). "Abnormal centrosome amplification in the absence of p53". Science 271 (5256): 1744–1747. doi:10.1126/science.271.5256.1744. PMID 8596939
- ^ Meraldi, P.; Honda, R.; Nigg, E.A. (2002). "Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53–/– cells". EMBO J 21 (4): 483–492. doi:10.1093/emboj/21.4.483. PMC 125866. PMID 11847097. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=125866
- ^ Storchova, Z.; Pellman, D. (2004). "From polyploidy to aneuploidy, genome instability and cancer". Nat Rev Mol Cell Biol. 5 (1): 45–54. doi:10.1038/nrm1276. PMID 14708009
- ^ Salisbury, J. L.; Suino, K. M.; Busby, R.; Springett, M. (2002). "Centrin-2 is required for centriole duplication in mammalian cells". Current biology : CB 12 (15): 1287–1292. doi:10.1016/S0960-9822(02)01019-9. PMID 12176356.
- ^ a b Marshall, W. F. (2009). "Centriole evolution". Current Opinion in Cell Biology 21 (1): 14–15. doi:10.1016/j.ceb.2009.01.008. PMC 2835302. PMID 19196504. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2835302.
- ^ Alliegro, M. C.; Alliegro, M. A.; Palazzo, R. E. (2006). "Centrosome-associated RNA in surf clam oocytes". Proceedings of the National Academy of Sciences 103 (24): 9034–9038. Bibcode 2006PNAS..103.9034A. doi:10.1073/pnas.0602859103. PMC 1482561. PMID 16754862. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1482561.
The centrosome and its components Centrioles Pericentriolar material other proteins B strc: edmb (perx), skel (ctrs), epit, cili, mito, nucl (chro) Structures of the cell / organelles (TH H1.00.01.2-3) Endomembrane system Cytoskeleton Endosymbionts Other internal External B strc: edmb (perx), skel (ctrs), epit, cili, mito, nucl (chro) Categories:- Organelles
Wikimedia Foundation. 2010.