- Multivariate normal distribution
-
"MVN" redirects here. For the airport with that IATA code, see Mount Vernon Airport.
Probability density function
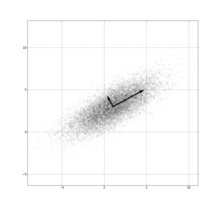
Many samples from a multivariate (bivariate) Gaussian distribution centered at (1,3) with a standard deviation of 3 in roughly the (0.878, 0.478) direction and of 1 in the orthogonal direction.notation: 
parameters: μ ∈ Rk — location
Σ ∈ Rk×k — covariance (nonnegative-definite matrix)support: x ∈ μ+span(Σ) ⊆ Rk pdf: 
exists only when Σ is positive-definitecdf: (no analytic expression) mean: μ mode: μ variance: Σ entropy: 
mgf: 
cf: 
In probability theory and statistics, the multivariate normal distribution or multivariate Gaussian distribution, is a generalization of the one-dimensional (univariate) normal distribution to higher dimensions. A random vector is said to be multivariate normally distributed if every linear combination of its components has a univariate normal distribution. The multivariate normal distribution is often used to describe, at least approximately, any set of (possibly) correlated real-valued random variables each of which clusters around a mean value.
Notation and parametrization
The multivariate normal distribution of a k-dimensional random vector X = [X1, X2, …, Xk] can be written in the following notation:
or to make it explicitly known that X is k-dimensional,
with k-dimensional mean vector
and k x k covariance matrix
Definition
A random vector X = (X1, …, Xk)′ is said to have the multivariate normal distribution if it satisfies the following equivalent conditions.[1]
- Every linear combination of its components Y = a1X1 + … + akXk is normally distributed. That is, for any constant vector a ∈ Rk, the random variable Y = a′X has a univariate normal distribution.
- There exists a random ℓ-vector Z, whose components are independent standard normal random variables, a k-vector μ, and a k×ℓ matrix A, such that X = AZ + μ. Here ℓ is the rank of the covariance matrix Σ = AA′. Especially in the case of full rank, see the section below on Geometric Interpretation.
- There is a k-vector μ and a symmetric, nonnegative-definite k×k matrix Σ, such that the characteristic function of X is
The covariance matrix is allowed to be singular (in which case the corresponding distribution has no density). This case arises frequently in statistics; for example, in the distribution of the vector of residuals in the ordinary least squares regression. Note also that the Xi are in general not independent; they can be seen as the result of applying the matrix A to a collection of independent Gaussian variables Z.
Properties
Density function
Non-degenerate case
The multivariate normal distribution is said to be "non-degenerate" when the covariance matrix Σ of the multivariate normal distribution is symmetric and positive definite. In this case the distribution has density
where
 is the determinant of A. Note how the equation above reduces to that of the univariate normal distribution if Σ is a
is the determinant of A. Note how the equation above reduces to that of the univariate normal distribution if Σ is a  matrix (ie a real number).
matrix (ie a real number).Bivariate case
In the 2-dimensional nonsingular case (k = rank(Σ) = 2), the probability density function of a vector [X Y]′ is
where ρ is the correlation between X and Y. In this case,
In the bivariate case, we also have a theorem that makes the first equivalent condition for multivariate normality less restrictive: it is sufficient to verify that countably many distinct linear combinations of X and Y are normal in order to conclude that the vector [X Y]′ is bivariate normal.[2]
Degenerate case
If the covariance matrix Σ is not full rank, then the multivariate normal distribution is degenerate and does not have a density. More precisely, it does not have a density with respect to k-dimensional Lebesgue measure (which is the usual measure assumed in calculus-level probability courses). Only random vectors whose distributions are absolutely continuous with respect to a measure are said to have densities (with respect to that measure). To talk about densities but avoid dealing with measure-theoretic complications it can be simpler to restrict attention to a subset of rank(Σ) of the coordinates of X such that covariance matrix for this subset is positive definite; then the other coordinates may be thought of as an affine function of the selected coordinates.
To talk about densities meaningfully in the singular case, then, we must select a different base measure. Using the disintegration theorem we can define a restriction of Lebesgue measure to the rank(Σ)-dimensional affine subspace of
 where the Gaussian distribution is supported, i.e.
where the Gaussian distribution is supported, i.e.  . With respect to this probability measure the distribution has density:
. With respect to this probability measure the distribution has density:where Σ − is the generalized inverse and det* is the pseudo-determinant.
Likelihood function
If the mean and variance matrix are unknown, a suitable log likelihood function for a single observation 'x' would be:

where x is a vector of real numbers. The complex case would be
 .
. Cumulative distribution function
The cumulative distribution function (cdf) F(x) of a random vector X is defined as the probability that all components of X are less than or equal to the corresponding values in the vector x. Though there is no closed form for F(x), there are a number of algorithms that estimate it numerically.
Normally distributed and independent
If X and Y are normally distributed and independent, this implies they are "jointly normally distributed", i.e., the pair (X, Y) must have multivariate normal distribution. However, a pair of jointly normally distributed variables need not be independent.
Two normally distributed random variables need not be jointly bivariate normal
The fact that two random variables X and Y both have a normal distribution does not imply that the pair (X, Y) has a joint normal distribution. A simple example is one in which X has a normal distribution with expected value 0 and variance 1, and Y = X if |X| > c and Y = −X if |X| < c, where c > 0. There are similar counterexamples for more than two random variables.[citation needed]
Conditional distributions
If μ and Σ are partitioned as follows
 with sizes
with sizes 
 with sizes
with sizes 
then, the distribution of X1 conditional on X2 = a is multivariate normal (X1|X2 = a) ∼ N(μ, Σ) where
and covariance matrix
This matrix is the Schur complement of Σ22 in Σ. This means that to calculate the conditional covariance matrix, one inverts the overall covariance matrix, drops the rows and columns corresponding to the variables being conditioned upon, and then inverts back to get the conditional covariance matrix. Here
 is the generalized inverse of Σ22
is the generalized inverse of Σ22
Note that knowing that X2 = a alters the variance, though the new variance does not depend on the specific value of a; perhaps more surprisingly, the mean is shifted by ; compare this with the situation of not knowing the value of a, in which case X1 would have distribution
; compare this with the situation of not knowing the value of a, in which case X1 would have distribution  .
.An interesting fact derived in order to prove this result, is that the random vectors X2 and
 are independent.
are independent.The matrix Σ12Σ22− is known as the matrix of regression coefficients.
In the bivariate case the conditional distribution of Y given X is
where ρ is the correlation coefficient between X and Y.
Bivariate conditional expectation
In the case
the following result holds[citation needed]
where the final ratio here is called the inverse Mills ratio.
Marginal distributions
To obtain the marginal distribution over a subset of multivariate normal random variables, one only needs to drop the irrelevant variables (the variables that one wants to marginalize out) from the mean vector and the covariance matrix. The proof for this follows from the definitions of multivariate normal distributions and linear algebra.[4]
Example
Let X = [X1, X2, X3] be multivariate normal random variables with mean vector μ = [μ1μ2μ3] and covariance matrix Σ (Standard parametrization for multivariate normal distribution). Then the joint distribution of X′ = [X1X3] is multivariate normal with mean vector μ′ = [μ1μ3] and covariance matrix
 .
.Affine transformation
If Y = c + BX is an affine transformation of
 where c is an
where c is an  vector of constants and B is a constant
vector of constants and B is a constant  matrix, then Y has a multivariate normal distribution with expected value c + Bμ and variance BΣBT i.e.,
matrix, then Y has a multivariate normal distribution with expected value c + Bμ and variance BΣBT i.e.,  . In particular, any subset of the Xi has a marginal distribution that is also multivariate normal. To see this, consider the following example: to extract the subset (X1, X2, X4)T, use
. In particular, any subset of the Xi has a marginal distribution that is also multivariate normal. To see this, consider the following example: to extract the subset (X1, X2, X4)T, usewhich extracts the desired elements directly.
Another corollary is that the distribution of Z = b · X, where b is a constant vector of the same length as X and the dot indicates a vector product, is univariate Gaussian with
 . This result follows by using
. This result follows by usingand considering only the first component of the product (the first row of B is the vector b). Observe how the positive-definiteness of Σ implies that the variance of the dot product must be positive.
An affine transformation of X such as 2X is not the same as the sum of two independent realisations of X.
Geometric interpretation
The equidensity contours of a non-singular multivariate normal distribution are ellipsoids (i.e. linear transformations of hyperspheres) centered at the mean.[5] The directions of the principal axes of the ellipsoids are given by the eigenvectors of the covariance matrix Σ. The squared relative lengths of the principal axes are given by the corresponding eigenvalues.
If Σ = UΛUT = UΛ1/2(UΛ1/2)T is an eigendecomposition where the columns of U are unit eigenvectors and Λ is a diagonal matrix of the eigenvalues, then we have
Moreover, U can be chosen to be a rotation matrix, as inverting an axis does not have any effect on N(0, Λ), but inverting a column changes the sign of U's determinant. The distribution N(μ, Σ) is in effect N(0, I) scaled by Λ1/2, rotated by U and translated by μ.
Conversely, any choice of μ, full rank matrix U, and positive diagonal entries Λi yields a non-singular multivariate normal distribution. If any Λi is zero and U is square, the resulting covariance matrix UΛUT is singular. Geometrically this means that every contour ellipsoid is infinitely thin and has zero volume in n-dimensional space, as at least one of the principal axes has length of zero.
Correlations and independence
In general, random variables may be uncorrelated but highly dependent. But if a random vector has a multivariate normal distribution then any two or more of its components that are uncorrelated are independent. This implies that any two or more of its components that are pairwise independent are independent.
But it is not true that two random variables that are (separately, marginally) normally distributed and uncorrelated are independent. Two random variables that are normally distributed may fail to be jointly normally distributed, i.e., the vector whose components they are may fail to have a multivariate normal distribution. For an example of two normally distributed random variables that are uncorrelated but not independent, see normally distributed and uncorrelated does not imply independent.
Higher moments
Main article: Isserlis’ theoremThe kth-order moments of X are defined by
where r1 + r2 + ⋯ + rN = k.
The central k-order central moments are given as follows
(a) If k is odd, μ1, …, N(X − μ) = 0.
(b) If k is even with k = 2λ, then
where the sum is taken over all allocations of the set
 into λ (unordered) pairs. That is, if you have a kth ( = 2λ = 6) central moment, you will be summing the products of λ = 3 covariances (the -μ notation has been dropped in the interests of parsimony):
into λ (unordered) pairs. That is, if you have a kth ( = 2λ = 6) central moment, you will be summing the products of λ = 3 covariances (the -μ notation has been dropped in the interests of parsimony):This yields (2λ − 1)! / (2λ − 1(λ − 1)!) terms in the sum (15 in the above case), each being the product of λ (in this case 3) covariances. For fourth order moments (four variables) there are three terms. For sixth-order moments there are 3 × 5 = 15 terms, and for eighth-order moments there are 3 × 5 × 7 = 105 terms.
The covariances are then determined by replacing the terms of the list
![\left[ 1,\dots,2\lambda \right]](8/218c7d67059fcf04a548d845d9f6e1d7.png) by the corresponding terms of the list consisting of r1 ones, then r2 twos, etc.. To illustrate this, examine the following 4th-order central moment case:
by the corresponding terms of the list consisting of r1 ones, then r2 twos, etc.. To illustrate this, examine the following 4th-order central moment case:where σij is the covariance of Xi and Xj. The idea with the above method is you first find the general case for a kth moment where you have k different X variables -
![E\left[ X_i X_j X_k X_n\right]](7/cf7446bc0444c0d68bd8dd68dd68a195.png) and then you can simplify this accordingly. Say, you have
and then you can simplify this accordingly. Say, you have ![E\left[ X_i^2 X_k X_n\right]](f/bdf61709456f398f3ff0535c90a2da35.png) then you simply let Xi = Xj and realise that σii = σi2.
then you simply let Xi = Xj and realise that σii = σi2.Kullback–Leibler divergence
The Kullback–Leibler divergence from
 to
to  , for non-singular matrices Σ0 and Σ1, is:
, for non-singular matrices Σ0 and Σ1, is:The logarithm must be taken to base e since the two terms following the logarithm are themselves base-e logarithms of expressions that are either factors of the density function or otherwise arise naturally. The equation therefore gives a result measured in nats. Dividing the entire expression above by loge 2 yields the divergence in bits.
Estimation of parameters
The derivation of the maximum-likelihood estimator of the covariance matrix of a multivariate normal distribution is perhaps surprisingly subtle and elegant. See estimation of covariance matrices.
In short, the probability density function (pdf) of an N-dimensional multivariate normal is
and the ML estimator of the covariance matrix from a sample of n observations is
which is simply the sample covariance matrix. This is a biased estimator whose expectation is
An unbiased sample covariance is
The Fisher information matrix for estimating the parameters of a multivariate normal distribution has a closed form expression. This can be used, for example, to compute the Cramér–Rao bound for parameter estimation in this setting. See Fisher information for more details.
Entropy
The differential entropy of the multivariate normal distribution is [7]
where
 is the determinant of the covariance matrix Σ.
is the determinant of the covariance matrix Σ.Multivariate normality tests
Multivariate normality tests check a given set of data for similarity to the multivariate normal distribution. The null hypothesis is that the data set is similar to the normal distribution, therefore a sufficiently small p-value indicates non-normal data. Multivariate normality tests include the Cox-Small test [8] and Smith and Jain's adaptation [9] of the Friedman-Rafsky test.[10]
Mardia's test[11] is based on multivariate extensions of skewness and kurtosis measures. For a sample {x1, ..., xn} of p-dimensional vectors we compute
Under the null hypothesis of multivariate normality, the statistic A will have approximately a chi-squared distribution with 1 6 ⋅p(p + 1)(p + 2) degrees of freedom, and B will be approximately standard normal N(0,1).
Mardia's kurtosis statistic is skewed and converges very slowly to the limiting normal distribution. For medium size samples
 , the parameters of the asymptotic distribution of the kurtosis statistic are modified [12] For small sample tests (n < 50) empirical critical values are used. Tables of critical values for both statistics are given by Rencher[13] for d=2,3,4.
, the parameters of the asymptotic distribution of the kurtosis statistic are modified [12] For small sample tests (n < 50) empirical critical values are used. Tables of critical values for both statistics are given by Rencher[13] for d=2,3,4.Mardia's tests are affine invariant but not consistent. For example, the multivariate skewness test is not consistent against symmetric non-normal alternatives.
The BHEP test[14] computes the norm of the difference between the empirical characteristic function and the theoretical characteristic function of the normal distribution. Calculation of the norm is performed in the L2(μ) space of square-integrable functions with respect to the Gaussian weighting function
 . The test statistic is
. The test statistic isThe limiting distribution of this test statistic is a weighted sum of chi-squared random variables, however in practice it is more convenient to compute the sample quantiles using the Monte-Carlo simulations.
A detailed survey of these and other test procedures is given by Henze (2002).
Drawing values from the distribution
A widely used method for drawing a random vector X from the N-dimensional multivariate normal distribution with mean vector μ and covariance matrix Σ works as follows:
- Find any real matrix A such that A AT = Σ. When Σ is positive-definite, the Cholesky decomposition is typically used. In the more general nonnegative-definite case, one can use the matrix A = UΛ½ obtained from a spectral decomposition Σ = UΛUT of Σ.
- Let Z = (z1, …, zN)T be a vector whose components are N independent standard normal variates (which can be generated, for example, by using the Box–Muller transform).
- Let X be μ + AZ. This has the desired distribution due to the affine transformation property.
See also
- Chi distribution, the pdf of the 2-norm (or Euclidean norm) of a multivariate normally-distributed vector.
- Complex normal distribution, for the generalization to complex valued random variables.
- Multivariate stable distribution extension of the multivariate normal distribution, when the index (exponent in the characteristic function) is between zero to two.
- Mahalanobis distance
- Wishart distribution
References
- ^ Gut, Allan: An Intermediate Course in Probability, 2009, chapter 5
- ^ Hamedani & Tata (1975)
- ^ Eaton, Morris L. (1983). Multivariate Statistics: a Vector Space Approach. John Wiley and Sons. pp. 116-117. ISBN 0-471-02776-6.
- ^ The formal proof for marginal distribution is shown here http://fourier.eng.hmc.edu/e161/lectures/gaussianprocess/node7.html
- ^ Nikolaus Hansen. "The CMA Evolution Strategy: A Tutorial" (PDF). http://www.bionik.tu-berlin.de/user/niko/cmatutorial.pdf.
- ^ Penny & Roberts, PARG-00-12, (2000) [1]. pp. 18
- ^ Gokhale, DV; NA Ahmed, BC Res, NJ Piscataway (May 1989). "Entropy Expressions and Their Estimators for Multivariate Distributions". Information Theory, IEEE Transactions on 35 (3): 688–692. doi:10.1109/18.30996.
- ^ Cox, D. R.; N. J. H. Small (August 1978). "Testing multivariate normality". Biometrika 65 (2): 263–272. doi:10.1093/biomet/65.2.263.
- ^ Smith, Stephen P.; Anil K. Jain (September 1988). "A test to determine the multivariate normality of a dataset". IEEE Transactions on Pattern Analysis and Machine Intelligence 10 (5): 757–761. doi:10.1109/34.6789.
- ^ Friedman, J. H. and Rafsky, L. C. (1979) "Multivariate generalizations of the Wald-Wolfowitz and Smirnov two sample tests". Annals of Statistics, 7, 697–717.
- ^ Mardia (1970)
- ^ Rencher (1995), pages 112-113.
- ^ Rencher (1995), pages 493-495.
- ^ First suggested by Epps & Pulley (1983), the consistency and limiting distribution derived by Baringhaus & Henze (1988).
Literature
- Baringhaus, L.; Henze, N. (1988). "A consistent test for multivariate normality based on the empirical characteristic function". Metrika 35 (1): 339–348. doi:10.1007/BF02613322.
- Epps, Lawrence B.; Pulley, Lawrence B. (1983). "A test for normality based on the empirical characteristic function". Biometrika 70 (3): 723–726. doi:10.1093/biomet/70.3.723.
- Hamedani, G. G.; Tata, M. N. (1975). "On the determination of the bivariate normal distribution from distributions of linear combinations of the variables". The American Mathematical Monthly 82 (9): 913–915. doi:10.2307/2318494.
- Henze, Norbert (2002). "Invariant tests for multivariate normality: a critical review". Statistical papers 43 (4): 467–506. doi:10.1007/s00362-002-0119-6.
- Mardia, K. V. (1970). "Measures of multivariate skewness and kurtosis with applications". Biometrika 57 (3): 519–530. doi:10.1093/biomet/57.3.519.
- Rencher, A.C. (1995). Methods of Multivariate Analysis. New York: Wiley.
Categories:- Continuous distributions
- Multivariate continuous distributions
- Normal distribution
Wikimedia Foundation. 2010.



![\mu = [ \operatorname{E}[X_1], \operatorname{E}[X_2], \ldots, \operatorname{E}[X_k]]](6/656df1a05805f7ac9e1509b61697acb7.png)
![\Sigma = [\operatorname{Cov}[X_i, X_j]], i=1,2,\ldots,k; j=1,2,\ldots,k](1/37116f4349eb02fdab11a01790a5ad82.png)


![f(x,y) =
\frac{1}{2 \pi \sigma_x \sigma_y \sqrt{1-\rho^2}}
\exp\left(
-\frac{1}{2(1-\rho^2)}\left[
\frac{(x-\mu_x)^2}{\sigma_x^2} +
\frac{(y-\mu_y)^2}{\sigma_y^2} -
\frac{2\rho(x-\mu_x)(y-\mu_y)}{\sigma_x \sigma_y}
\right]
\right),](5/e359479df2f05b3125526ede1ad68cf4.png)










![\mu _{1,\dots,N}(X)\ \stackrel{\mathrm{def}}{=}\ \mu _{r_{1},\dots,r_{N}}(X)\ \stackrel{\mathrm{def}}{=}\ E\left[
\prod\limits_{j=1}^{N}X_j^{r_{j}}\right]](8/8e895d7ed390749881b8992984ae3fa8.png)

![\begin{align}
& {} E[X_1 X_2 X_3 X_4 X_5 X_6] \\
&{} = E[X_1 X_2 ]E[X_3 X_4 ]E[X_5 X_6 ] + E[X_1 X_2 ]E[X_3 X_5 ]E[X_4 X_6] + E[X_1 X_2 ]E[X_3 X_6 ]E[X_4 X_5] \\
&{} + E[X_1 X_3 ]E[X_2 X_4 ]E[X_5 X_6 ] + E[X_1 X_3 ]E[X_2 X_5 ]E[X_4 X_6 ] + E[X_1 X_3]E[X_2 X_6]E[X_4 X_5] \\
&+ E[X_1 X_4]E[X_2 X_3]E[X_5 X_6]+E[X_1 X_4]E[X_2 X_5]E[X_3 X_6]+E[X_1 X_4]E[X_2 X_6]E[X_3 X_5] \\
& + E[X_1 X_5]E[X_2 X_3]E[X_4 X_6]+E[X_1 X_5]E[X_2 X_4]E[X_3 X_6]+E[X_1 X_5]E[X_2 X_6]E[X_3 X_4] \\
& + E[X_1 X_6]E[X_2 X_3]E[X_4 X_5 ] + E[X_1 X_6]E[X_2 X_4 ]E[X_3 X_5] + E[X_1 X_6]E[X_2 X_5]E[X_3 X_4].
\end{align}](7/477220038abb3f6d2ee172e33bcde13f.png)
![E\left[ X_i^4\right] = 3\sigma _{ii}^2](e/7ee572fbf3fbe2077e2f886d7710d1d4.png)
![E\left[ X_i^3 X_j\right] = 3\sigma _{ii} \sigma _{ij}](7/bd7a84adc118f6fef0d3e44fd515f707.png)
![E\left[ X_i^2 X_j^2\right] = \sigma _{ii}\sigma_{jj}+2\left( \sigma _{ij}\right) ^2](3/4b34e16082722ca5e14bc525aa275597.png)
![E\left[ X_i^2X_jX_k\right] = \sigma _{ii}\sigma _{jk}+2\sigma _{ij}\sigma _{ik}](5/ea55493c283fd864bcbe46c5da9c4ee5.png)
![E\left[ X_i X_j X_k X_n\right] = \sigma _{ij}\sigma _{kn}+\sigma _{ik}\sigma _{jn}+\sigma _{in}\sigma _{jk}.](6/67659b5bf0652636330890c6b45712d7.png)



![E[\widehat\Sigma] = {n-1 \over n}\Sigma.](5/6f5b2fd1317b7743d57df74a3e9350dc.png)


![\begin{align}
& \hat\Sigma = \frac{1}{n} \sum_{j=1}^n (x_j - \bar x)(x_j - \bar x)' \\
& A = \frac{1}{6n} \sum_{i=1}^n \sum_{j=1}^n \Big[ (x_i - \bar x)'\hat\Sigma^{-1} (x_j - \bar x) \Big]^3 \\
& B = \frac{\sqrt{n}}{\sqrt{8p(p+2)}}\bigg[\frac{1}{n} \sum_{i=1}^n \Big[ (x_i - \bar x)'\hat\Sigma^{-1} (x_i - \bar x) \Big]^2 - p(p+2) \bigg]
\end{align}](c/29c778262dc11512ca11b6f7451a7100.png)
