- L-form bacteria
-
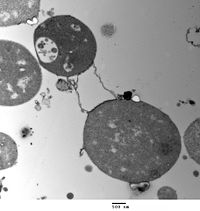 Transmission electron micrograph of L-form Bacillus subtilis. The cells lack the electron-dense cell wall of normal bacteria. Scale bar is 500 nanometers.
Transmission electron micrograph of L-form Bacillus subtilis. The cells lack the electron-dense cell wall of normal bacteria. Scale bar is 500 nanometers.
L-form bacteria, also known as L-phase bacteria, L-phase variants, and cell wall-deficient (CWD) bacteria, are strains of bacteria that lack cell walls.[1] They were first isolated in 1935 by Emmy Klieneberger-Nobel, who named them "L-forms" after the Lister Institute in London where she was working[2].
Two types of L-forms are distinguished: unstable L-forms, spheroplasts that are capable of dividing, but can revert to the original morphology, and stable L-forms, L-forms that are unable to revert to the original bacteria.
Some parasitic species of bacteria, such as mycoplasma, also lack a cell wall[3], but these are not considered L-forms since they are not derived from bacteria that normally have cell walls[4].
Contents
Appearance and cell division
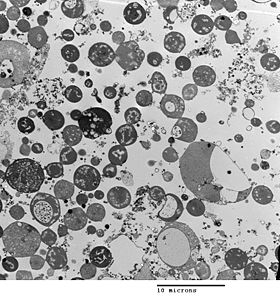 Transmission electron micrograph of L-form Bacillus subtilis, showing a range of sizes. Scale bar is 10 micrometers.
Transmission electron micrograph of L-form Bacillus subtilis, showing a range of sizes. Scale bar is 10 micrometers.
Bacterial morphology is determined by the cell wall. As L-form has no cell wall, its morphology is different from that of the strain of bacteria from which it is derived. Typical L-form cells are spheres or spheroids. For example, L-forms of the rod-shaped bacterium Bacillus subtilis appear round when viewed by phase contrast microscopy or by transmission electron microscopy.[5]
Although L-forms can develop from Gram-positive as well as from Gram-negative bacteria, in a Gram stain test, the L-forms always colour Gram-negative, due to the lack of a cell wall.
The cell wall is important for cell division, which, in most bacteria, occurs by binary fission. The lack of cell wall in L-forms means that division is disorganised, giving rise to a variety of cell sizes, from very tiny to very big.
 Phase contrast image of L-form cells from Bacillus subtilis showing a range of sizes. Scale bar is 5 micrometers.
Phase contrast image of L-form cells from Bacillus subtilis showing a range of sizes. Scale bar is 5 micrometers.
In bacteria, cell division usually requires a cell wall and components of the bacterial cytoskeleton such as FtsZ. The ability of L-form bacteria to grow and divide in the absence of both of these structures is highly unusual, and may represent a form of cell division that was important in early forms of life.[1] This novel mode of division seems to involve the extension of thin protrusions from the cell's surface and these protrusions then pinching off to form new cells.
Generation in cultures
L-forms can be generated in the laboratory from many bacterial species that usually have cell walls, such as Bacillus subtilis or Escherichia coli. This is done by inhibiting peptidoglycan synthesis with antibiotics or treating the cells with lysozyme, an enzyme that digests cell walls. The L-forms are generated in a culture medium that is the same osmolarity as the bacterial cytosol (an isotonic solution), which prevents cell lysis by osmotic shock.[2] L-form strains can be unstable, tending to revert to the normal form of the bacteria by regrowing a cell wall, but this can be prevented by long-term culture of the cells under the same conditions that were used to produce them.[6]. Some studies have identified mutations that occur, as these strains are derived from normal bacteria.[1][2] One such point mutation is in an enzyme involved in the mevalonate pathway of lipid metabolism that increased the frequency of L-form formation 1,000-fold.[1] The reason for this effect is not known, but it is presumed that the increase is related to this enzyme's role in making a lipid important in peptidoglycan synthesis.
Another methodology of induction relies on nanotechnology and landscape ecology. Microfluidics devices can be built in order to challenge peptidoglycan synthesis by extreme spatial confinement. After biological dispersal through a constricted (sub-micrometre scale) biological corridor connecting adjacent micro habitat patches, L-form-like cells can be derived [7].
Significance and applications
Laboratory-generated L-form strains have not been proven to exist in nature. Some publications have argued that L-form bacteria might cause human diseases, but, as the evidence that links these organisms to disease is fragmentary and frequently contradictory, this hypothesis remains controversial.[8][9] The two extreme viewpoints on this question are that L-form bacteria are either laboratory curiosities of no clinical significance or important but unappreciated causes of disease.[4] Research on L-form bacteria is continuing. For example, L-form organisms have been observed in mouse lungs after experimental inoculation with Nocardia caviae,[10][11] and a recent study suggested that these organisms may infect immunosuppressed patients having undergone bone marrow transplants.[12] The formation of strains of bacteria lacking cell walls has also been proposed to be important in the acquisition of bacterial antibiotic resistance.[13]
L-form bacteria may be useful in research on early forms of life, and in biotechnology. These strains are being examined for possible uses in biotechnology as host strains for recombinant protein expression.[14] Here, the absence of a cell wall can allow production of large amounts of secreted proteins that would otherwise accumulate in the periplasmic space of bacteria.[15][16] The accumulation of proteins in the periplasmic space can be toxic to the bacteria, which may reduce the yield of the expressed proteins.
See also
- Protoplast
- Spheroplast
- Mycoplasmataceae — lack peptidoglycan, but possess a mycolic acid layer for stability
- Ultramicrobacteria
Further reading
- Domingue, Gerald J. (1982). Cell wall-deficient bacteria: basic principles and clinical significance. Reading, Mass: Addison-Wesley Pub. Co. ISBN 0-201-10162-9.
- Mattman, Lida H. (2001). Cell wall deficient forms: stealth pathogens. Boca Raton: CRC. ISBN 0-8493-8767-1. http://books.google.com/?id=mincr2Hi81UC&pg=frontpage.
External links
- Errington Group at Newcastle University
- Scientists explore new window on the origins of life 2009 Newcastle University press release
References
- ^ a b c d Leaver M, Domínguez-Cuevas P, Coxhead JM, Daniel RA, Errington J (February 2009). "Life without a wall or division machine in Bacillus subtilis". Nature 457 (7231): 849–53. doi:10.1038/nature07742. PMID 19212404. http://www.nature.com/nature/journal/v457/n7231/full/nature07742.html.
- ^ a b c Joseleau-Petit D, Liébart JC, Ayala JA, D'Ari R (September 2007). "Unstable Escherichia coli L Forms Revisited: Growth Requires Peptidoglycan Synthesis". J. Bacteriol. 189 (18): 6512–20. doi:10.1128/JB.00273-07. PMC 2045188. PMID 17586646. http://jb.asm.org/cgi/pmidlookup?view=long&pmid=17586646.
- ^ Razin S, Yogev D, Naot Y (December 1998). "Molecular Biology and Pathogenicity of Mycoplasmas". Microbiol. Mol. Biol. Rev. 62 (4): 1094–156. PMC 98941. PMID 9841667. http://mmbr.asm.org/cgi/pmidlookup?view=long&pmid=9841667.
- ^ a b Domingue GJ, Woody HB (April 1997). "Bacterial persistence and expression of disease". Clin. Microbiol. Rev. 10 (2): 320–44. PMC 172922. PMID 9105757. http://cmr.asm.org/cgi/pmidlookup?view=long&pmid=9105757. Full PDF
- ^ Gilpin RW, Young FE, Chatterjee AN (January 1973). "Characterization of a Stable L-Form of Bacillus subtilis 168". J. Bacteriol. 113 (1): 486–99. PMC 251652. PMID 4631836. http://jb.asm.org/cgi/pmidlookup?view=long&pmid=4631836.
- ^ Allan EJ (April 1991). "Induction and cultivation of a stable L-form of Bacillus subtilis". J. Appl. Bacteriol. 70 (4): 339–43. doi:10.1111/j.1365-2672.1991.tb02946.x. PMID 1905284.
- ^ Männik J., R. Driessen, P. Galajda, J.E. Keymer, C. Dekker (September 2009). "Bacterial growth and motility in sub-micron constrictions". PNAS 106 (35): 14861–14866. doi:10.1073/pnas.0907542106. PMC 2729279. PMID 19706420. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2729279.
- ^ Onwuamaegbu ME, Belcher RA, Soare C (2005). "Cell wall-deficient bacteria as a cause of infections: a review of the clinical significance". J. Int. Med. Res. 33 (1): 1–20. PMID 15651712. http://www.jimronline.net/content/full/2005/58/0545.pdf.
- ^ Casadesús J (December 2007). "Bacterial L-forms require peptidoglycan synthesis for cell division". Bioessays 29 (12): 1189–91. doi:10.1002/bies.20680. PMID 18008373.
- ^ Beaman BL (July 1980). "Induction of L-phase variants of Nocardia caviae within intact murine lungs". Infect. Immun. 29 (1): 244–51. PMC 551102. PMID 7399704. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=551102.
- ^ Beaman BL, Scates SM (September 1981). "Role of L-forms of Nocardia caviae in the development of chronic mycetomas in normal and immunodeficient murine models". Infect. Immun. 33 (3): 893–907. PMC 350795. PMID 7287189. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=350795.
- ^ Woo PC, Wong SS, Lum PN, Hui WT, Yuen KY (March 2001). "Cell-wall-deficient bacteria and culture-negative febrile episodes in bone-marrow-transplant recipients". Lancet 357 (9257): 675–9. doi:10.1016/S0140-6736(00)04131-3. PMID 11247551.
- ^ Fuller E, Elmer C, Nattress F, et al (December 2005). "β-Lactam Resistance in Staphylococcus aureus Cells That Do Not Require a Cell Wall for Integrity". Antimicrob. Agents Chemother. 49 (12): 5075–80. doi:10.1128/AAC.49.12.5075-5080.2005. PMC 1315936. PMID 16304175. http://aac.asm.org/cgi/content/full/49/12/5075#R10.
- ^ Gumpert J, Hoischen C (October 1998). "Use of cell wall-less bacteria (L-forms) for efficient expression and secretion of heterologous gene products". Curr. Opin. Biotechnol. 9 (5): 506–9. doi:10.1016/S0958-1669(98)80037-2. PMID 9821280.
- ^ Rippmann JF, Klein M, Hoischen C, et al (1 December 1998). "Procaryotic Expression of Single-Chain Variable-Fragment (scFv) Antibodies: Secretion in L-Form Cells of Proteus mirabilis Leads to Active Product and Overcomes the Limitations of Periplasmic Expression in Escherichia coli". Appl. Environ. Microbiol. 64 (12): 4862–9. PMC 90935. PMID 9835575. http://aem.asm.org/cgi/pmidlookup?view=long&pmid=9835575.
- ^ Choi JH, Lee SY (June 2004). "Secretory and extracellular production of recombinant proteins using Escherichia coli". Appl. Microbiol. Biotechnol. 64 (5): 625–35. doi:10.1007/s00253-004-1559-9. PMID 14966662.
Pathogenic bacteria Bacterial disease · Coley's Toxins · Exotoxin · Lysogenic cycle
Human flora Substrate preference Oxygen preference Structures Cell wall: Peptidoglycan (NAM, NAG, DAP)
Gram-positive bacteria only: Teichoic acid · Lipoteichoic acid · Endospore
Gram-negative bacteria only: Bacterial outer membrane (Porin, Lipopolysaccharide) · Periplasmic space
Mycobacteria only: Arabinogalactan · Mycolic acidOutside envelopeCompositeShapes Categories:
Wikimedia Foundation. 2010.
