- Nucleic acid tertiary structure
-
 Example of a large catalytic RNA. The self-splicing group II intron from Oceanobacillus iheyensis.[1]
Example of a large catalytic RNA. The self-splicing group II intron from Oceanobacillus iheyensis.[1]
The tertiary structure of a nucleic acid is its precise three-dimensional structure, as defined by the atomic coordinates.[2] RNA and DNA molecules are capable of diverse functions ranging from molecular recognition to catalysis. Such functions require a precise three-dimensional tertiary structure. While such structures are diverse and seemingly complex, they are composed of recurring, easily recognizable tertiary structure motifs that serve as molecular building blocks. Some of the most common motifs for RNA and DNA tertiary structure are described below, but it is important to remember that this information is based on a limited number of solved structures. Many more tertiary structural motifs will be revealed as new RNA and DNA molecules are structurally characterized.
Contents
Helical structures
Double helix
Main article: Nucleic acid double helixThe double helix is the dominant tertiary structure for biological DNA, and is also a possible structure for RNA. Three DNA conformations are believed to be found in nature, A-DNA, B-DNA, and Z-DNA. The "B" form described by James D. Watson and Francis Crick is believed to predominate in cells.[3] James D. Watson and Francis Crick described this structure as a double helix with a radius of 10 Å and pitch of 34 Å, making one complete turn about its axis every 10 bp of sequence.[4] The double helix makes one complete turn about its axis every 10.4-10.5 base pairs in solution. This frequency of twist (known as the helical pitch) depends largely on stacking forces that each base exerts on its neighbours in the chain. Double-helical RNA adopts a conformation similar to the A-form structure.
Other conformations are possible; in fact, only the letters F, Q, U, V, and Y are now available to describe any new DNA structure that may appear in the future.[5][6] However, most of these forms have been created synthetically and have not been observed in naturally occurring biological systems.
RNA triplexes
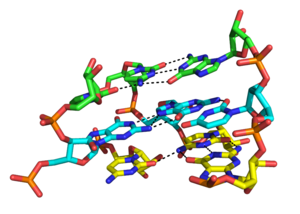
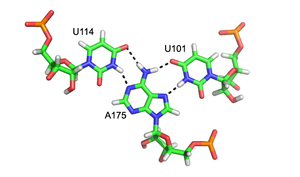
Major groove triples in the group II intron in Oceanobacillus Iheyensis. Each stacked layer is formed by one triplex with a different color scheme. Hydrogen bonds between triplexes are shown in black dashed lines. "N" atoms are colored in blue and "O" atoms in red. From top to bottom, the residues on the left side are G288, C289, and C377.[7]Close-up rendering of the U114:A175-U101 major groove (Hoogsteen base) triplex formed within the wild type pseudoknot of Human Telomerase RNA. Hydrogen bonds are shown in black dashed lines. "N" atoms are colored in blue and "o" atoms in red.[8]Major and minor groove triplexes
The minor groove triple is a ubiquitous RNA structural motif. Because interactions with the minor groove are often mediated by the 2'-OH of the ribose sugar, this RNA motif looks very different from its DNA equivalent. The most common example of a minor loop triple is the A-minor motif, or the insertion of adenosine bases into the minor groove (see above). However, this motif is not restricted to adenosines, as other nucleobases have also been observed to interact with the RNA minor groove.
The minor groove presents a near-perfect complement for an inserted base. This allows for optimal van der Waals contacts, extensive hydrogen bonding and hydrophobic surface burial, and creates a highly energetically favorable interaction.[9] Because minor groove triples are capable of stably packing a free loop and helix, they are key elements in the structure of large ribonucleotides, including the group I intron,[10] the group II intron,[11] and the ribosome.
Although the major groove of standard A-form RNA is fairly narrow and therefore less available for triplex interaction than the minor groove, major groove triplex interactions can be observed in several RNA structures. These structures consist of several combinations of base pair and Hoogsteen interactions. For example, the GGC triplex (GGC amino(N-2)-N-7, imino-carbonyl, carbonyl-amino(N-4); Watson-Crick) observed in the 50S ribosome, composed of a Watson-Crick type G-C pair and an incoming G which forms a pseudo-Hoogsteen network of hydrogen bonding interactions between both bases involved in the canonical pairing.[12] Other notable examples of major groove triplexes include (i) the catalytic core of the group II intron shown in the figure at left [7] (ii) a catalytically essential triple helix observed in human telomerase RNA[8] and (iii) the SAM-II riboswitch.[14]
Triple-stranded DNA is also possible from Hoogsteen or reversed Hoogsteen hydrogen bonds in the major groove of B-form DNA.
Quadruplexes
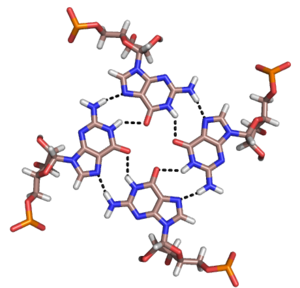
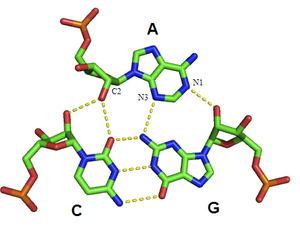
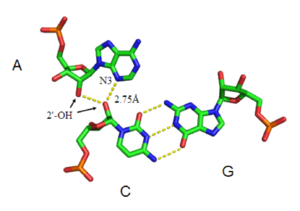
Besides double helices and the above-mentioned triplexes, RNA and DNA can both also form quadruple helices. There are diverse structures of RNA base quadruplexes. Four consecutive guanine residues can form a quadruplex in RNA by Hoogsteen hydrogen bonds to form a “Hoogsteen ring” (See Figure).[12] G-C and A-U pairs can also form base quadruplex with a combination of Watson-Crick pairing and noncanonical pairing in the minor groove.[15]
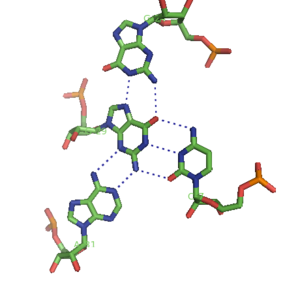
The core of malachite green aptamer is also a kind of base quadruplex with a different hydrogen bonding pattern (See Figure).[13] The quadruplex can repeat several times consecutively, producing an immensely stable structure.
The unique structure of quadruplex regions in RNA may serve different functions in a biological system. Two important functions are the binding potential with ligands or proteins, and its ability to stabilize the whole tertiary structure of DNA or RNA. The strong structure can inhibit or modulate transcription and replication, such as in the telomeres of chromosomes and the UTR of mRNA.[16] The base identity is important towards ligand binding. The G-quartet typically binds monovalent cations such as potassium, while other bases can bind numerous other ligands such as hypoxanthine in a U-U-C-U quadruplex.[15]
Along with these functions, the G-quadruplex in the mRNA around the ribosome binding regions could serve as a regulator of gene expression in bacteria.[17] There may be more interesting structures and functions yet to be discovered in vivo.
Coaxial stacking
 Secondary (inset) and tertiary structure of tRNA demonstrating coaxial stacking.[18]
Secondary (inset) and tertiary structure of tRNA demonstrating coaxial stacking.[18]
Coaxial stacking, otherwise known as helical stacking, is a major determinant of higher order RNA tertiary structure. Coaxial stacking occurs when two RNA duplexes form a contiguous helix, which is stabilized by base stacking at the interface of the two helices. Coaxial stacking was noted in the crystal structure of tRNAPhe.[19] More recently, coaxial stacking has been observed in higher order structures of many ribozymes, including many forms of the self-splicing group I and group II introns. Common coaxial stacking motifs include the kissing loop interaction and the pseudoknot. The stability of these interactions can be predicted by an adaptation of “Turner’s rules”.[20]
In 1994, Walter and Turner determined the free energy contributions of nearest neighbor stacking interactions within a helix-helix interface by using a model system that created a helix-helix interface between a short oligomer and a four-nucleotide overhang at the end of a hairpin stem . Their experiments confirmed that the thermodynamic contribution of base-stacking between two helical secondary structures closely mimics the thermodynamics of standard duplex formation (nearest neighbor interactions predict the thermodynamic stability of the resulting helix). The relative stability of nearest neighbor interactions can be used to predict favorable coaxial stacking based on known secondary structure. Walter and Turner found that, on average, prediction of RNA structure improved from 67% to 74% accuracy when coaxial stacking contributions were included.[21] Theories of coaxial stacking can be tested using the technique of helical fusion. This approach was used by Murphy and Cech to confirm a coaxial stacking interaction between the P4 and P6 helices within the catalytic center of the Tetrahymena group I intron.
Most well-studied RNA tertiary structures contain examples of coaxial stacking. Some prominent examples are tRNA-Phe, group I introns, group II introns, and ribosomal RNAs. Crystal structures of tRNA revealed the presence of two extended helices that result from coaxial stacking of the amino-acid acceptor stem with the T-arm, and stacking of the D- and anticodon-arms. These interactions within tRNA orient the anticodon stem perpendicularly to the amino-acid stem, leading to the functional L-shaped tertiary structure.[19] In group I introns, the P4 and P6 helices were shown to coaxially stack using a combination of biochemical[22] and crystallographic methods. The P456 crystal structure provided a detailed view of how coaxial stacking stabilizes the packing of RNA helices into tertiary structures.[23] In the self-splicing group II intron from Oceanobacillus iheyensis, the IA and IB stems coaxially stack and contribute to the relative orientation of the constituent helices of a five-way junction.[7] This orientation facilitates proper folding of the active site of the functional ribozyme. The ribosome contains numerous examples of coaxial stacking, including stacked segments as long as 70 bp.[24]
Two common motifs involving coaxial stacking are kissing loops and pseudoknots. In kissing loop interactions, the single-stranded loop regions of two hairpins interact through base pairing, forming a composite, coaxially stacked helix. Notably, this structure allows all of the nucleotides in each loop to participate in base-pairing and stacking interactions. This motif was visualized and studied using NMR analysis by Lee and Crothers.[25] The pseudoknot motif occurs when a single stranded region of a hairpin loop basepairs with an upstream or downstream sequence within the same RNA strand. The two resulting duplex regions often stack upon one another, forming a stable coaxially stacked composite helix. One example of a pseudoknot motif is the highly stable Hepatitis Delta virus ribozyme, in which the backbone shows an overall double pseudoknot topology.[26]
An effect similar to coaxial stacking has been observed in rationally designed DNA structures. DNA origami structures contain a large number of double helixes with exposed blunt ends. These structures were observed to stick together along the edges that contained these exposed blunt ends, due to the hydrophobic stacking interactions.[27]
Other motifs
Tetraloop-receptor interactions
 Stick representation of a GAAA tetraloop - an example from the GNRA tetraloop family.[28]
Stick representation of a GAAA tetraloop - an example from the GNRA tetraloop family.[28]
Tetraloop-receptor interactions combine base-pairing and stacking interactions between the loop nucleotides of a tetraloop motif and a receptor motif located within an RNA duplex, creating a tertiary contact that stabilizes the global tertiary fold of an RNA molecule. Tetraloops are also possible structures in DNA duplexes.[29]
Stem-loops can vary greatly in size and sequence, but tetraloops of four nucleotides are very common and they usually belong to one of three categories, based on sequence.[30] These three families are the CUYG, UNCG and GNRA (see figure on the right) tetraloops.[31] In each of these tetraloop families, the second and third nucleotides form a turn in the RNA strand and a base-pair between the first and fourth nucleotides stabilizes the stemloop structure. It has been determined, in general, that the stability of the tetraloop depends on the composition of bases within the loop and on the composition of this "closing base pair".[32] The GNRA family of tetraloops is the most commonly observed within Tetraloop-receptor interactions.
 GAAA Tetraloop and Receptor: Stick representation of tetraloop (yellow) and its receptor, showing both Watson-Crick and Hoogsteen base-pairing.[28]
GAAA Tetraloop and Receptor: Stick representation of tetraloop (yellow) and its receptor, showing both Watson-Crick and Hoogsteen base-pairing.[28]
“Tetraloop receptor motifs” are long-range tertiary interactions[33] consisting of hydrogen bonding between the bases in the tetraloop to stemloop sequences in distal sections of the secondary RNA structure.[34] In addition to hydrogen bonding, stacking interactions are an important component of these tertiary interactions. For example, in GNRA-tetraloop interactions, the second nucleotide of the tetraloop stacks directly on an A-platform motif (see above) within the receptor.[23] The sequence of the tetraloop and its receptor often covary so that the same type of tertiary contact can be made with different isoforms of the tetraloop and its cognate receptor.[35]
For example, the self-splicing group I intron relies on tetraloop receptor motifs for its structure and function.[23][34] Specifically, the three adenine residues of the canonical GAAA motif stack on top of the receptor helix and form multiple stabilizing hydrogen bonds with the receptor. The first adenine of the GAAA sequence forms a triple base-pair with the receptor AU bases. The second adenine is stabilized by hydrogen bonds with the same uridine, as well as via its 2'-OH with the recptor and via interactions with the guanine of the GAAA tetraloop. The third adenine forms a triple base pair.
A-minor motif
A-minor Interactions
Type I A-minor interaction: Type I interactions are the most common, strongest A-minor interactions, as they involve numerous hydrogen bonds, and bury the incoming A base in the minor groove.[36]The A-minor motif is a ubiquitous RNA tertiary structural motif. It is formed by the insertion of an unpaired nucleoside into the minor groove of an RNA duplex. As such it is an example of a minor groove triple. Although guanosine, cytosine and uridine can also form minor groove triple interactions, minor groove interactions by adenine are very common. In the case of adenine, the N1-C2-N3 edge of the inserting base forms hydrogen bonds with one or both of the 2’-OH’s of the duplex, as well as the bases of the duplex (see figure: A-minor interactions). The host duplex is often a G-C basepair.
A-minor motifs have been separated into four classes,[9] types 0 to III, based upon the position of the inserting base relative to the two 2’-OH’s of the Watson-Crick base pair. In type I and II A-minor motifs, N3 of adenine is inserted deeply within the minor groove of the duplex (see figure: A minor interactions - type II interaction), and there is good shape complementarity with the base pair. Unlike types 0 and III, type I and II interactions are specific for adenine due to hydrogen bonding interactions. In the type III interaction, both the O2' and N3 of the inserting base are associated less closely with the minor groove of the duplex. Type 0 and III motifs are weaker and non-specific because they are mediated by interactions with a single 2’-OH (see figure: A-minor Interactions - type 0 and type III interactions).
The A-minor motif is among the most common RNA structural motifs in the ribosome, where it contributes to the binding of tRNA to the 23S subunit.[37] They most often stabilize RNA duplex interactions in loops and helices, such as in the core of group II introns.[7]
An interesting example of A-minor is its role in anticodon recognition. The ribosome must discriminate between correct and incorrect codon-anticodon pairs. It does so, in part, through the insertion of adenine bases into the minor groove. Incorrect codon-anticodon pairs will present distorted helical geometry, which will prevent the A-minor interaction from stabilizing the binding, and increase the dissociation rate of the incorrect tRNA.[38]
Ribose zipper
 Ribose Zippers: View of a canonical ribose zipper between two RNA backbones.[28]
Ribose Zippers: View of a canonical ribose zipper between two RNA backbones.[28]
The ribose zipper is an RNA tertiary structural element in which two RNA chains are held together by hydrogen bonding interactions involving the 2’OH of ribose sugars on different strands. The 2'OH can behave as both hydrogen bond donor and acceptor, which allows formation of bifurcated hydrogen bonds with another 2’ OH.[39][40]
Numerous forms of ribose zipper have been reported, but a common type involves four hydrogen bonds between 2'-OH groups of two adjacent sugars. Ribose zippers commonly occur in arrays that stabilize interactions between separate RNA strands.[1] Ribose zippers are often observed as Stem-loop interactions with very low sequence specificity. However, in the small and large ribosomal subunits, there exists a propensity for ribose zippers of the CC/AA sequence- two cytosines on the first chain paired to two adenines on the second chain.
Role of metal ions
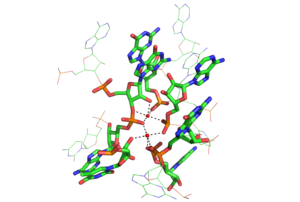
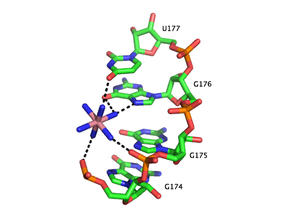
Metal Ion Binding in the Group I Intron
PDB rendering of Group I intron inner sphere magnesium coordination. The two red balls indicate magnesium ions and dashed lines coming from the ions indicate coordination with the respective groups on nucleotides. The color-coding scheme is as follows: green=carbon, orange=phosphate, pink=oxygen, blue=nitrogen.[41]PDB rendering of Group 1 intron P5c binding pocket demonstrating outer sphere coordination. Here, the six amines of osmium hexamine(III) fulfill the role generally served by water molecules and mediate the ion’s interaction with the major groove. Coordination via hydrogen bonds is indicated by dashed lines and osmium is rendered in pink, all other colors are as above.[28]Functional RNAs are often folded, stable molecules with three-dimensional shapes rather than floppy, linear strands.[42] Cations are essential for thermodynamic stabilization of RNA tertiary structures. Metal cations that bind RNA can be monovalent, divalent or trivalent. Potassium (K+) is a common monovalent ion that binds RNA. A common divalent ion that binds RNA is magnesium (Mg2+). Other ions including sodium (Na+), calcium (Ca2+) and manganese (Mn2+) have been found to bind RNA in vivo and in vitro. Multivalent organic cations such as spermidine or spermine are also found in cells and these make important contributions to RNA folding. Trivalent ions such as cobalt hexamine or lanthanide ions such as terbium (Tb3+) are useful experimental tools for studying metal binding to RNA.[43]
A metal ion can interact with RNA in multiple ways. An ion can associate diffusely with the RNA backbone, shielding otherwise unfavorable electrostatic interactions. This charge screening is often fulfilled by monovalent ions. Site-bound ions stabilize specific elements of RNA tertiary structure. Site-bound interactions can be further subdivided into two categories depending on whether water mediates the metal binding. “Outer sphere” interactions are mediated by water molecules that surround the metal ion. For example, magnesium hexahydrate interacts with and stabilizes specific RNA tertiary structure motifs via interactions with guanosine in the major groove. Conversely, “inner sphere” interactions are directly mediated by the metal ion. RNA often folds in multiple stages and these steps can be stabilized by different types of cations. In the early stages, RNA forms secondary structures stabilized through the binding of monovalent cations, divalent cations and polyanionic amines in order to neutralize the polyanionic backbone. The later stages of this process involve the formation of RNA tertiary structure, which is stabilized almost largely through the binding of divalent ions such as magnesium with possible contributions from potassium binding.
Metal-binding sites are often localized in the deep and narrow major groove of the RNA duplex, coordinating to the Hoogsteen edges of purines. In particular, metal cations stabilize sites of backbone twisting where tight packing of phosphates results in a region of dense negative charge. There are several metal ion-binding motifs in RNA duplexes that have been identified in crystal structures. For instance, in the P4-P6 domain of the Tetrahymena thermophila group I intron, several ion-binding sites consist of tandem G-U wobble pairs and tandem G-A mismatches, in which divalent cations interact with the Hoogsteen edge of guanosine via O6 and N7.[44][45][46] Another ion-binding motif in the Tetrahymena group I intron is the A-A platform motif, in which consecutive adenosines in the same strand of RNA form a non-canonical pseudobase pair.[47] Unlike the tandem G-U motif, the A-A platform motif binds preferentially to monovalent cations. In many of these motifs, absence of the monovalent or divalent cations results in either greater flexibility or loss of tertiary structure.
Divalent metal ions, especially magnesium, have been found to be important for the structure of DNA junctions such as the Holliday junction intermediate in genetic recombination. The magnesium ion shields the negatively-charged phosphate groups in the junction and allows them to be positioned closer together, allowing a stacked conformation rather than an unstacked conformation.[48] Magnesium is vital in stabilizing these kinds of junctions in artificially designed structures used in DNA nanotechnology, such as the double crossover motif.[49]
History
Main article: History of molecular biologyThe earliest work in RNA structural biology coincided, more or less, with the work being done on DNA in the early 1950s. In their seminal 1953 paper, Watson and Crick suggested that van der Waals crowding by the 2`OH group of ribose would preclude RNA from adopting a double helical structure identical to the model they proposed - what we now know as B-form DNA.[50] This provoked questions about the three dimensional structure of RNA: could this molecule form some type of helical structure, and if so, how?
In the mid 1960's, the role of tRNA in protein synthesis was being intensively studied. In 1965, Holley et al. purified and sequenced the first tRNA molecule, initially proposing that it adopted a cloverleaf structure, based largely on the ability of certain regions of the molecule to form stem loop structures.[51] The isolation of tRNA proved to be the first major windfall in RNA structural biology. In 1971, Kim et al. achieved another breakthrough, producing crystals of yeast tRNAPHE that diffracted to 2-3 Ångström resolutions by using spermine, a naturally occurring polyamine, which bound to and stabilized the tRNA.[52]
For a considerable time following the first tRNA structures, the field of RNA structure did not dramatically advance. The ability to study an RNA structure depended upon the potential to isolate the RNA target. This proved limiting to the field for many years, in part owing to the fact that other known targets - i.e. the ribosome - were significantly more difficult to isolate and crystallize. As such, for some twenty years following the original publication of the tRNAPHE structure, the structures of only a handful of other RNA targets were solved, with almost all of these belonging to the transfer RNA family.[53]
This unfortunate lack of scope would eventually be overcome largely because of two major advancements in nucleic acid research: the identification of ribozymes, and the ability to produce them via in vitro transcription. Subsequent to Tom Cech's publication implicating the Tetrahymena group I intron as an autocatalytic ribozyme,[54] and Sidney Altman's report of catalysis by ribonuclease P RNA,[55] several other catalytic RNAs were identified in the late 1980s,[56] including the hammerhead ribozyme. In 1994, McKay et al. published the structure of a 'hammerhead RNA-DNA ribozyme-inhibitor complex' at 2.6 Ångström resolution, in which the autocatalytic activity of the ribozyme was disrupted via binding to a DNA substrate.[57] In addition to the advances being made in global structure determination via crystallography, the early 1990s also saw the implementation of NMR as a powerful technique in RNA structural biology. Investigations such as this enabled a more precise characterization of the base pairing and base stacking interactions which stabilized the global folds of large RNA molecules.
The resurgence of RNA structural biology in the mid-1990s has caused a veritable explosion in the field of nucleic acid structural research. Since the publication of the hammerhead and P4-6 structures, numerous major contributions to the field have been made. Some of the most noteworthy examples include the structures of the Group I and Group II introns,[7] and the Ribosome.[36] It should be noted that the first three structures were produced using in vitro transcription, and that NMR has played a role in investigating partial components of all four structures - testaments to the indispensability of both techniques for RNA research. Most recently, the 2009 Nobel Prize in Chemistry was awarded to Ada Yonath, Venkatraman Ramakrishnan, and Thomas Steitz for their structural work on the ribosome, demonstrating the prominent role RNA structural biology has taken in modern molecular biology.
See also
References
- ^ a b PDB 3IGI; Toor N, Keating KS, Fedorova O, Rajashankar K, Wang J, Pyle AM (January 2010). "Tertiary architecture of the Oceanobacillus iheyensis group II intron". RNA 16 (1): 57–69. doi:10.1261/rna.1844010. PMC 2802037. PMID 19952115. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2802037.; rendered using PyMOL.
- ^ IUPAC, Compendium of Chemical Terminology, 2nd ed. (the "Gold Book") (1997). Online corrected version: (2006–) "tertiary structure".
- ^ Richmond et al. (2003). "The structure of DNA in the nucleosome core". Nature 423 (6936): 145–150. doi:10.1038/nature01595. PMID 12736678.
- ^ Watson J.D. and Crick F.H.C. (1953). "A Structure for Deoxyribose Nucleic Acid" (PDF). Nature 171 (4356): 737–738. Bibcode 1953Natur.171..737W. doi:10.1038/171737a0. PMID 13054692. http://www.nature.com/nature/dna50/watsoncrick.pdf.
- ^ Bansal M (2003). "DNA structure: Revisiting the Watson-Crick double helix". Current Science 85 (11): 1556–1563.
- ^ Ghosh A, Bansal M (2003). "A glossary of DNA structures from A to Z". Acta Cryst D59 (4): 620–626. doi:10.1107/S0907444903003251. PMID 12657780.
- ^ a b c d e PDB 3BWP; Toor N, Keating KS, Taylor SD, Pyle AM (April 2008). "Crystal structure of a self-spliced group II intron". Science 320 (5872): 77–82. doi:10.1126/science.1153803. PMID 18388288.; rendered with PyMOL
- ^ a b PDB 2K95; Kim NK, Zhang Q, Zhou J, Theimer CA, Peterson RD, Feigon J (December 2008). "Solution structure and dynamics of the wild-type pseudoknot of human telomerase RNA". J. Mol. Biol. 384 (5): 1249–61. doi:10.1016/j.jmb.2008.10.005. PMC 2660571. PMID 18950640. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2660571.; rendered with PyMOL
- ^ a b Doherty EA, Batey RT, Masquida B, Doudna JA (April 2001). "A universal mode of helix packing in RNA". Nat. Struct. Biol. 8 (4): 339–43. doi:10.1038/86221. PMID 11276255.
- ^ Szewczak AA, Ortoleva-Donnelly L, Ryder SP, Moncoeur E, Strobel SA (December 1998). "A minor groove RNA triple helix within the catalytic core of a group I intron". Nat. Struct. Biol. 5 (12): 1037–42. doi:10.1038/4146. PMID 9846872.
- ^ Boudvillain M, de Lencastre A, Pyle AM (July 2000). "A tertiary interaction that links active-site domains to the 5' splice site of a group II intron". Nature 406 (6793): 315–8. doi:10.1038/35018589. PMID 10917534.
- ^ a b c PDB 1RAU; Cheong C, Moore PB (September 1992). "Solution structure of an unusually stable RNA tetraplex containing G- and U-quartet structures". Biochemistry 31 (36): 8406–14. doi:10.1021/bi00151a003. PMID 1382577.; rendered with PyMOL
- ^ a b PDB 1FIT; Baugh C, Grate D, Wilson C (August 2000). "2.8 A crystal structure of the malachite green aptamer". J. Mol. Biol. 301 (1): 117–28. doi:10.1006/jmbi.2000.3951. PMID 10926496.; rendered with PyMOL
- ^ Gilbert SD, Rambo RP, Van Tyne D, Batey RT (February 2008). "Structure of the SAM-II riboswitch bound to S-adenosylmethionine". Nat. Struct. Mol. Biol. 15 (2): 177–82. doi:10.1038/nsmb.1371. PMID 18204466.
- ^ a b Batey RT, Gilbert SD, Montange RK (November 2004). "Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine". Nature 432 (7015): 411–5. doi:10.1038/nature03037. PMID 15549109.
- ^ Arthanari H, Bolton PH (March 2001). "Functional and dysfunctional roles of quadruplex DNA in cells". Chem. Biol. 8 (3): 221–30. doi:10.1016/S1074-5521(01)00007-2. PMID 11306347.
- ^ Oliver AW, Bogdarina I, Schroeder E, Taylor IA, Kneale GG (August 2000). "Preferential binding of fd gene 5 protein to tetraplex nucleic acid structures". J. Mol. Biol. 301 (3): 575–84. doi:10.1006/jmbi.2000.3991. PMID 10966771.
- ^ PDB 6tna; Sussman JL, Holbrook SR, Warrant RW, Church GM, Kim SH (August 1978). "Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement". J. Mol. Biol. 123 (4): 607–30. doi:10.1016/0022-2836(78)90209-7. PMID 357742.; rendered via PyMOL.
- ^ a b Quigley GJ, Rich A (November 1976). "Structural domains of transfer RNA molecules". Science 194 (4267): 796–806. doi:10.1126/science.790568. PMID 790568.
- ^ "Douglas H. Turner". Turner’s rules. Department of Chemistry, University of Rochester. http://www.chem.rochester.edu/faculty/faculty.php?name=turner.
- ^ Walter AE, Turner DH, Kim J, Lyttle MH, Müller P, Mathews DH, Zuker M (September 1994). "Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding". Proc. Natl. Acad. Sci. U.S.A. 91 (20): 9218–22. doi:10.1073/pnas.91.20.9218. PMC 44783. PMID 7524072. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=44783.
- ^ Murphy FL, Wang YH, Griffith JD, Cech TR (September 1994). "Coaxially stacked RNA helices in the catalytic center of the Tetrahymena ribozyme". Science 265 (5179): 1709–12. doi:10.1126/science.8085157. PMID 8085157.
- ^ a b c Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA (September 1996). "Crystal structure of a group I ribozyme domain: principles of RNA packing". Science 273 (5282): 1678–85. doi:10.1126/science.273.5282.1678. PMID 8781224.
- ^ Noller HF (September 2005). "RNA structure: reading the ribosome". Science 309 (5740): 1508–14. doi:10.1126/science.1111771. PMID 16141058.
- ^ Lee AJ, Crothers DM (August 1998). "The solution structure of an RNA loop-loop complex: the ColE1 inverted loop sequence". Structure 6 (8): 993–1005. doi:10.1016/S0969-2126(98)00101-4. PMID 9739090.
- ^ Ferré-D'Amaré AR, Zhou K, Doudna JA (October 1998). "Crystal structure of a hepatitis delta virus ribozyme". Nature 395 (6702): 567–74. doi:10.1038/26912. PMID 9783582.
- ^ Rothemund, Paul W. K. (2006). "Folding DNA to create nanoscale shapes and patterns". Nature 440 (7082): 297–302. doi:10.1038/nature04586. ISSN 0028-0836. PMID 16541064.
- ^ a b c d PDB 1GID; Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA (September 1996). "Crystal structure of a group I ribozyme domain: principles of RNA packing". Science 273 (5282): 1678–85. doi:10.1126/science.273.5282.1678. PMID 8781224.; rendered with PyMOL
- ^ Nakano, M; Moody, EM; Liang, J; Bevilacqua, PC (2002). "Selection for thermodynamically stable DNA tetraloops using temperature gradient gel electrophoresis reveals four motifs: d(cGNNAg), d(cGNABg),d(cCNNGg), and d(gCNNGc)". Biochemistry 41 (48): 14281–92. doi:10.1021/bi026479k. PMID 12450393.
- ^ Moore PB (1999). "Structural motifs in RNA". Annu. Rev. Biochem. 68 (1): 287–300. doi:10.1146/annurev.biochem.68.1.287. PMID 10872451.
- ^ Abramovitz DL, Pyle AM (February 1997). "Remarkable morphological variability of a common RNA folding motif: the GNRA tetraloop-receptor interaction". J. Mol. Biol. 266 (3): 493–506. doi:10.1006/jmbi.1996.0810. PMID 9067606.
- ^ Moody EM, Feerrar JC, Bevilacqua PC (June 2004). "Evidence that folding of an RNA tetraloop hairpin is less cooperative than its DNA counterpart". Biochemistry 43 (25): 7992–8. doi:10.1021/bi049350e. PMID 15209494.
- ^ Williams DH, Gait MJ, Loakes D (2006). Nucleic Acids in Chemistry and Biology. Cambridge, UK: RSC Pub. ISBN 0-85404-654-2.
- ^ a b Jaeger L, Michel F, Westhof E (March 1994). "Involvement of a GNRA tetraloop in long-range RNA tertiary interactions". J. Mol. Biol. 236 (5): 1271–6. doi:10.1016/0022-2836(94)90055-8. PMID 7510342.
- ^ Michel F, Westhof E (December 1990). "Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis". J. Mol. Biol. 216 (3): 585–610. doi:10.1016/0022-2836(90)90386-Z. PMID 2258934.
- ^ a b c PDB 1FFK; Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (August 2000). "The complete atomic structure of the large ribosomal subunit at 2.4 A resolution". Science 289 (5481): 905–20. doi:10.1126/science.289.5481.905. PMID 10937989.; rendered with PyMOL
- ^ Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA (April 2001). "RNA tertiary interactions in the large ribosomal subunit: the A-minor motif". Proc. Natl. Acad. Sci. U.S.A. 98 (9): 4899–903. doi:10.1073/pnas.081082398. PMC 33135. PMID 11296253. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=33135.
- ^ Yoshizawa S, Fourmy D, Puglisi JD (September 1999). "Recognition of the codon-anticodon helix by ribosomal RNA". Science 285 (5434): 1722–5. doi:10.1126/science.285.5434.1722. PMID 10481006.
- ^ Batey RT, Rambo RP, Doudna JA (August 1999). "Tertiary Motifs in RNA Structure and Folding". Angew. Chem. Int. Ed. Engl. 38 (16): 2326–2343. doi:10.1002/(SICI)1521-3773(19990816)38:16<2326::AID-ANIE2326>3.0.CO;2-3. PMID 10458781.
- ^ Tamura M, Holbrook SR (July 2002). "Sequence and structural conservation in RNA ribose zippers". J. Mol. Biol. 320 (3): 455–74. doi:10.1016/S0022-2836(02)00515-6. PMID 12096903.
- ^ PDB 1ZZN; Stahley MR, Strobel SA (September 2005). "Structural evidence for a two-metal-ion mechanism of group I intron splicing". Science 309 (5740): 1587–90. doi:10.1126/science.1114994. PMID 16141079.; rendered with PyMOL
- ^ Celander DW, Cech TR (January 1991). "Visualizing the higher order folding of a catalytic RNA molecule". Science 251 (4992): 401–7. doi:10.1126/science.1989074. PMID 1989074.
- ^ Pyle AM (September 2002). "Metal ions in the structure and function of RNA". J. Biol. Inorg. Chem. 7 (7–8): 679–90. doi:10.1007/s00775-002-0387-6. PMID 12203005.
- ^ Cate JH, Doudna JA (October 1996). "Metal-binding sites in the major groove of a large ribozyme domain". Structure 4 (10): 1221–9. doi:10.1016/S0969-2126(96)00129-3. PMID 8939748.
- ^ Kieft JS, Tinoco I (May 1997). "Solution structure of a metal-binding site in the major groove of RNA complexed with cobalt (III) hexammine". Structure 5 (5): 713–21. doi:10.1016/S0969-2126(97)00225-6. PMID 9195889.
- ^ Rüdisser S, Tinoco I (February 2000). "Solution structure of Cobalt(III)hexammine complexed to the GAAA tetraloop, and metal-ion binding to G·A mismatches". J. Mol. Biol. 295 (5): 1211–23. doi:10.1006/jmbi.1999.3421. PMID 10653698.
- ^ Burkhardt C, Zacharias M (October 2001). "Modelling ion binding to AA platform motifs in RNA: a continuum solvent study including conformational adaptation". Nucleic Acids Res. 29 (19): 3910–8. PMC 60250. PMID 11574672. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=60250.
- ^ Panyutin, IG; Biswas, I; Hsieh, P (1995). "A pivotal role for the structure of the Holliday junction in DNA branch migration". The EMBO journal 14 (8): 1819–26. PMC 398275. PMID 7737132. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=398275.
- ^ Fu, Tsu Ju; Seeman, Nadrian C. (1993). "DNA double-crossover molecules". Biochemistry 32 (13): 3211–20. doi:10.1021/bi00064a003. PMID 8461289.
- ^ Watson JD, Crick FH (April 1953). "Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid" (PDF). Nature 171 (4356): 737–738. Bibcode 1953Natur.171..737W. doi:10.1038/171737a0. PMID 13054692. http://www.nature.com/nature/dna50/watsoncrick.pdf.
- ^ Holley, RW, Apgar, J, Everett, GA, Madison, JT, Marguisse, M, Merrill, SH, Penwick, JR, Zamir (March 1965). "Structure of a ribonucleic acid". Science 147 (3664): 1462–5. doi:10.1126/science.147.3664.1462. PMID 14263761.
- ^ Kim SH, Quigley G, Suddath FL, Rich A (April 1971). "High-resolution x-ray diffraction patterns of crystalline transfer RNA that show helical regions". Proc. Natl. Acad. Sci. U.S.A. 68 (4): 841–5. doi:10.1073/pnas.68.4.841. PMC 389056. PMID 5279525. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=389056.
- ^ Shen LX, Cai Z, Tinoco I (August 1995). "RNA structure at high resolution". FASEB J. 9 (11): 1023–33. PMID 7544309.
- ^ Cech TR, Zaug AJ, Grabowski PJ (December 1981). "In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence". Cell 27 (3 Pt 2): 487–96. doi:10.1016/0092-8674(81)90390-1. PMID 6101203.
- ^ Stark BC, Kole R, Bowman EJ, Altman S (August 1978). "Ribonuclease P: an enzyme with an essential RNA component". Proc. Natl. Acad. Sci. U.S.A. 75 (8): 3717–21. doi:10.1073/pnas.75.8.3717. PMC 392857. PMID 358197. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=392857.
- ^ Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G (March 1986). "Autolytic Processing of Dimeric Plant Virus Satellite RNA". Science 231 (4745): 1577–1580. doi:10.1126/science.231.4745.1577. PMID 17833317.
- ^ Pley HW, Flaherty KM, McKay DB (November 1994). "Three-dimensional structure of a hammerhead ribozyme". Nature 372 (6501): 68–74. doi:10.1038/372068a0. PMID 7969422.
Protein structure Nucleic acid structure See also Categories:
Wikimedia Foundation. 2010.