- Green fluorescent protein
-
"EGFP" redirects here. EGFP may also refer to the ICAO airport code for Pembrey Airport.
The green fluorescent protein (GFP) is a protein composed of 238 amino acid residues (26.9kDa) that exhibits bright green fluorescence when exposed to blue light.[1][2] Although many other marine organisms have similar green fluorescent proteins, GFP traditionally refers to the protein first isolated from the jellyfish Aequorea victoria. The GFP from A. victoria has a major excitation peak at a wavelength of 395 nm and a minor one at 475 nm. Its emission peak is at 509 nm, which is in the lower green portion of the visible spectrum. The fluorescence quantum yield (QY) of GFP is 0.79. The GFP from the sea pansy (Renilla reniformis) has a single major excitation peak at 498 nm. In cell and molecular biology, the GFP gene is frequently used as a reporter of expression.[3] In modified forms it has been used to make biosensors, and many animals have been created that express GFP as a proof-of-concept that a gene can be expressed throughout a given organism. The GFP gene can be introduced into organisms and maintained in their genome through breeding, injection with a viral vector, or cell transformation. To date, the GFP gene has been introduced and expressed in many bacteria, yeast and other fungi, fish (such as zebrafish), plant, fly, and mammalian cells, including human. Martin Chalfie, Osamu Shimomura, and Roger Y. Tsien were awarded the 2008 Nobel Prize in Chemistry on 10 October 2008 for their discovery and development of the green fluorescent protein.
Contents
History
Wild-type GFP (wtGFP)
In the 1960s and 1970s, GFP, along with the separate luminescent protein aequorin, was first purified from Aequorea victoria and its properties studied by Osamu Shimomura.[4] In A. victoria, GFP fluorescence occurs when aequorin interacts with Ca2+ ions, inducing a blue glow. Some of this luminescent energy is transferred to the GFP, shifting the overall color towards green.[5] However, its utility as a tool for molecular biologists did not begin to be realized until 1992 when Douglas Prasher reported the cloning and nucleotide sequence of wtGFP in Gene.[6] The funding for this project had run out, so Prasher sent cDNA samples to several labs. The lab of Martin Chalfie expressed the coding sequence of wtGFP, with the first few amino acids deleted, in heterologous cells of E. coli and C. elegans, publishing the results in Science in 1994.[7] Frederick Tsuji's lab independently reported the expression of the recombinant protein one month later.[8] Remarkably, the GFP molecule folded and was fluorescent at room temperature, without the need for exogenous cofactors specific to the jellyfish. Although this near-wtGFP was fluorescent, it had several drawbacks, including dual peaked excitation spectra, pH sensitivity, chloride sensitivity, poor fluorescence quantum yield, poor photostability and poor folding at 37°C.
The first reported crystal structure of a GFP was that of the S65T mutant by the Remington group in Science in 1996.[9] One month later, the Phillips group independently reported the wild-type GFP structure in Nature Biotech.[10] These crystal structures provided vital background on chromophore formation and neighboring residue interactions. Researchers have modified these residues by directed and random mutagenesis to produce the wide variety of GFP derivatives in use today. Martin Chalfie, Osamu Shimomura and Roger Y. Tsien share the 2008 Nobel Prize in Chemistry for their discovery and development of the green fluorescent protein.[11]
GFP derivatives
Due to the potential for widespread usage and the evolving needs of researchers, many different mutants of GFP have been engineered.[12] The first major improvement was a single point mutation (S65T) reported in 1995 in Nature by Roger Tsien.[13] This mutation dramatically improved the spectral characteristics of GFP, resulting in increased fluorescence, photostability, and a shift of the major excitation peak to 488 nm, with the peak emission kept at 509 nm. This matched the spectral characteristics of commonly available FITC filter sets, increasing the practicality of use by the general researcher. A 37 °C folding efficiency (F64L) point mutant to this scaffold yielding enhanced GFP (EGFP) was discovered in 1995 by the lab of Ole Thastrup.[14] EGFP allowed the practical use of GFPs in mammalian cells. EGFP has an extinction coefficient (denoted ε) of 55,000 M−1cm−1.[15] The fluorescence quantum yield (QY) of EGFP is 0.60. The relative brightness, expressed as ε•QY, is 33,000 M−1cm−1. Superfolder GFP, a series of mutations that allow GFP to rapidly fold and mature even when fused to poorly folding peptides, was reported in 2006.[16]
Many other mutations have been made, including color mutants; in particular, blue fluorescent protein (EBFP, EBFP2, Azurite, mKalama1), cyan fluorescent protein (ECFP, Cerulean, CyPet), and yellow fluorescent protein derivatives (YFP, Citrine, Venus, YPet). BFP derivatives (except mKalama1) contain the Y66H substitution. The critical mutation in cyan derivatives is the Y66W substitution, which causes the chromophore to form with an indole rather than phenol component. Several additional compensatory mutations in the surrounding barrel are required to restore brightness to this modified chromophore due to the increased bulk of the indole group. The red-shifted wavelength of the YFP derivatives is accomplished by the T203Y mutation and is due to π-electron stacking interactions between the substituted tyrosine residue and the chromophore.[2] These two classes of spectral variants are often employed for Förster resonance energy transfer (FRET) experiments. Genetically-encoded FRET reporters sensitive to cell signaling molecules, such as calcium or glutamate, protein phosphorylation state, protein complementation, receptor dimerization, and other processes provide highly specific optical readouts of cell activity in real time.
Semirational mutagenesis of a number of residues led to pH-sensitive mutants known as pHluorins, and later super-ecliptic pHluorins. By exploiting the rapid change in pH upon synaptic vesicle fusion, pHluorins tagged to synaptobrevin have been used to visualize synaptic activity in neurons.[17]
Redox sensitive versions of GFP (roGFP) were engineered by introduction of cysteines into the beta barrel structure. The redox state of the cysteines determines the fluorescent properties of roGFP.[18]
The nomenclature of modified GFPs is often confusing due to overlapping mapping of several GFP versions onto a single name. For example, mGFP often refers to a GFP with an N-terminal palmitoylation that causes the GFP to bind to cell membranes. However, the same term is also used to refer to monomeric GFP, which is often achieved by the dimer interface breaking A206K mutation.[19] Wild-type GFP has a weak dimerization tendency at concentrations above 5 mg/mL. mGFP also stands for "modified GFP," which has been optimized through amino acid exchange for stable expression in plant cells.
Structure
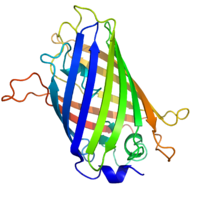
GFP has a typical beta barrel structure, consisting of one β-sheet with alpha helix(s) containing the covalently bonded chromophore 4-(p-hydroxybenzylidene)imidazolidin-5-one (HBI) running through the center.[2][9][10] HBI is nonfluorescent in the absence of the properly folded GFP scaffold and exists mainly in the unionized phenol form in wtGFP.[citation needed] Inward-facing sidechains of the barrel induce specific cyclization reactions in the tripeptide Ser65–Tyr66–Gly67 that induce ionization of HBI to the phenolate form and chromophore formation. This process of post-translational modification is referred to as maturation.[citation needed] The hydrogen-bonding network and electron-stacking interactions with these sidechains influence the color, intensity and photostability of GFP and its numerous derivatives.[citation needed] The tightly packed nature of the barrel excludes solvent molecules, protecting the chromophore fluorescence from quenching by water.
Green fluorescent protein 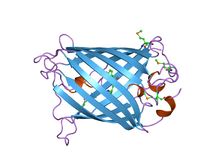
Structure of the Aequorea victoria green fluorescent protein.[20] Identifiers Symbol GFP Pfam PF01353 Pfam clan CL0069 InterPro IPR011584 SCOP 1ema Available protein structures: Pfam structures PDB RCSB PDB; PDBe PDBsum structure summary 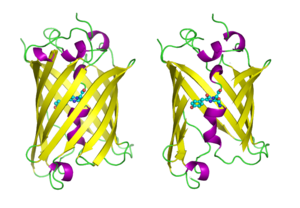 GFP molecules drawn in cartoon style, one fully and one with the side of the beta barrel cut away to reveal the chromophore (highlighted as ball-and-stick). From PDB 1GFL.
GFP molecules drawn in cartoon style, one fully and one with the side of the beta barrel cut away to reveal the chromophore (highlighted as ball-and-stick). From PDB 1GFL.
Use
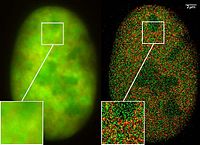
Superresolution with two fusion proteins (GFP-Snf2H and RFP-H2A), Co-localisation studies (2CLM) in the nucleus of a bone cancer cell. 120.000 localized molecules in a widefield area(470 µm2)
The availability of GFP and its derivatives has thoroughly redefined fluorescence microscopy and the way it is used in cell biology and other biological disciplines.[21] While most small fluorescent molecules such as FITC (fluorescein isothiocyanate) are strongly phototoxic when used in live cells, fluorescent proteins such as GFP are usually much less harmful when illuminated in living cells. This has triggered the development of highly automated live-cell fluorescence microscopy systems, which can be used to observe cells over time expressing one or more proteins tagged with fluorescent proteins. For example, GFP had been widely used in labelling the spermatozoa of various organisms for identification purposes as in Drosophila melanogaster, where expression of GFP can be used as a marker for a particular characteristic. GFP can also be expressed in different structures enabling morphological distinction. In such cases, the gene for the production of GFP is spliced into the genome of the organism in the region of the DNA that codes for the target proteins and that is controlled by the same regulatory sequence; that is, the gene's regulatory sequence now controls the production of GFP, in addition to the tagged protein(s). In cells where the gene is expressed, and the tagged proteins are produced, GFP is produced at the same time. Thus, only those cells in which the tagged gene is expressed, or the target proteins are produced, will fluoresce when observed under fluorescence microscopy. Analysis of such time lapse movies has redefined the understanding of many biological processes including protein folding, protein transport, and RNA dynamics, which in the past had been studied using fixed (i.e., dead) material.
The Vertico SMI microscope using the SPDM Phymod technology uses the so-called "reversible photobleaching" effect of fluorescent dyes like GFP and its derivatives to localize them as single molecules in an optical resolution of 10 nm. This can also be performed as a co-localization of two GFP derivatives (2CLM).[22]
Another powerful use of GFP is to express the protein in small sets of specific cells. This allows researchers to optically detect specific types of cells in vitro (in a dish), or even in vivo (in the living organism).[23] Genetically combining several spectral variants of GFP is a useful trick for the analysis of brain circuitry (Brainbow).[24] Other interesting uses of fluorescent proteins in the literature include using FPs as sensors of neuron membrane potential,[25] tracking of AMPA receptors on cell membranes,[26] viral entry and the infection of individual influenza viruses and lentiviral viruses,[27][28] etc.
It has also been found that new lines of transgenic GFP rats can be relevant for gene therapy as well as regenerative medicine.[29] By using "high-expresser" GFP, transgenic rats display high expression in most tissues, and many cells that have not been characterized or have been only poorly characterized in previous GFP-transgenic rats. Through its ability to form internal chromophore without requiring accessory cofactors, enzymes or substrates other than molecular oxygen, GFP makes for an excellent tool in all forms of biology.[30]
GFP has been shown to be useful in cryobiology as a viability assay. Correlation of viability as measured by trypan blue assays were 0.97.[31]
A novel possible use of GFP includes using it as a sensitive monitor of intracellular processes via an eGFP laser system made out of a human embryonic kidney cell line. The first engineered living laser is made by an eGFP expressing cell inside a reflective optical cavity and hitting it with pulses of blue light. At a certain pulse threshold, the eGFP’s optical output becomes brighter and completely uniform in color of pure green with a wavelength of 516 nm. Before being emitted as laser light, the light bounces back and forth within the resonator cavity and passes the cell numerous times. By studying the changes in optical activity, researchers may better understand cellular processes.[32][33]
GFP in nature
The purpose of both bioluminescence and GFP fluorescence in jellyfish is unknown. GFP is co-expressed with aequorin in small granules around the rim of the jellyfish bell. The secondary excitation peak (480 nm) of GFP does absorb some of the blue emission of aequorin, giving the bioluminescence a more green hue. The serine 65 residue of the GFP chromophore is responsible for the dual-peaked excitation spectra of wild-type GFP. It is conserved in all three GFP isoforms originally cloned by Prasher. Nearly all mutations of this residue consolidate the excitation spectra to a single peak at either 395 nm or 480 nm. The precise mechanism of this sensitivity is complex, but, it seems, involves donation of a hydrogen from serine 65 to glutamate 222, which influences chromophore ionization.[2] Since a single mutation can dramatically enhance the 480 nm excitation peak, making GFP a much more efficient partner of aequorin, A. victoria appears to evolutionarily prefer the less-efficient, dual-peaked excitation spectrum. Roger Tsien has speculated that varying hydrostatic pressure with depth may effect serine 65's ability to donate a hydrogen to the chromophore and shift the ratio of the two excitation peaks. Thus, the jellyfish may change the color of its bioluminescence with depth. However, a collapse in the population of jellyfish in Friday Harbor, where GFP was originally discovered, has hampered further study of the role of GFP in the jellyfish's natural environment.
GFP in fine art
 Julian Voss-Andreae's GFP-based sculpture Steel Jellyfish (2006). The image shows the stainless-steel sculpture at Friday Harbor Laboratories on San Juan Island (Wash., USA), the place of GFP's discovery.
Julian Voss-Andreae's GFP-based sculpture Steel Jellyfish (2006). The image shows the stainless-steel sculpture at Friday Harbor Laboratories on San Juan Island (Wash., USA), the place of GFP's discovery.
Julian Voss-Andreae, a German-born artist specializing in "protein sculptures,"[34] created sculptures based on the structure of GFP, including the 5'6" (1.70 m) tall "Green Fluorescent Protein" (2004)[35] and the 4'7" (1.40 m) tall "Steel Jellyfish" (2006). The latter sculpture is located at the place of GFP's discovery by Shimomura in 1962, the University of Washington's Friday Harbor Laboratories.[36]
Transgenic pets
Alba, a green-fluorescent rabbit, was created by a French laboratory commissioned by Eduardo Kac using GFP for purposes of art and social commentary.[37] The US company Yorktown Technologies markets to aquarium shops green fluorescent zebrafish (GloFish) that were initially developed to detect pollution in waterways. NeonPets, a US based company markets green fluorescent mice to the pet industry as NeonMice.[38] Green fluorescent pigs, known as Noels, were bred by a group of researchers led by Wu Shinn-Chih at the Department of Animal Science and Technology at National Taiwan University.[39] A Japanese-American Team created green-fluorescent cats as proof of concept to use them potentially as model organisms for diseases, particularly HIV.[40]
See also
References
- ^ Prendergast F, Mann K (1978). "Chemical and physical properties of aequorin and the green fluorescent protein isolated from Aequorea forskålea". Biochemistry 17 (17): 3448–53. doi:10.1021/bi00610a004. PMID 28749.
- ^ a b c d Tsien R (1998). "The green fluorescent protein" (PDF). Annu Rev Biochem 67: 509–44. doi:10.1146/annurev.biochem.67.1.509. PMID 9759496. http://tsienlab.ucsd.edu/Publications/Tsien%201998%20Annu.%20Rev.%20Biochem%20-%20GFP.pdf.
- ^ Phillips G (2001). "Green fluorescent protein--a bright idea for the study of bacterial protein localization". FEMS Microbiol Lett 204 (1): 9–18. doi:10.1016/S0378-1097(01)00358-5. PMID 11682170.
- ^ Shimomura O, Johnson F, Saiga Y (1962). "Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea". J Cell Comp Physiol 59 (3): 223–39. doi:10.1002/jcp.1030590302. PMID 13911999.
- ^ Morise H, Shimomura O, Johnson F, Winant J (1974). "Intermolecular energy transfer in the bioluminescent system of Aequorea". Biochemistry 13 (12): 2656–62. doi:10.1021/bi00709a028. PMID 4151620.
- ^ Prasher D, Eckenrode V, Ward W, Prendergast F, Cormier M (1992). "Primary structure of the Aequorea victoria green-fluorescent protein". Gene 111 (2): 229–33. doi:10.1016/0378-1119(92)90691-H. PMID 1347277.
- ^ Chalfie M, Tu Y, Euskirchen G, Ward W, Prasher D (1994). "Green fluorescent protein as a marker for gene expression". Science 263 (5148): 802–5. doi:10.1126/science.8303295. PMID 8303295.
- ^ Inouye S, Tsuji F (1994). "Aequorea green fluorescent protein. Expression of the gene and fluorescence characteristics of the recombinant protein". FEBS Lett 341 (2–3): 277–80. doi:10.1016/0014-5793(94)80472-9. PMID 8137953.
- ^ a b Ormö M, Cubitt A, Kallio K, Gross L, Tsien R, Remington S (1996). "Crystal structure of the Aequorea victoria green fluorescent protein". Science 273 (5280): 1392–5. doi:10.1126/science.273.5280.1392. PMID 8703075.
- ^ a b Yang F, Moss L, Phillips G (1996). "The molecular structure of green fluorescent protein". Nat Biotechnol 14 (10): 1246–51. doi:10.1038/nbt1096-1246. PMID 9631087.
- ^ "The Nobel Prize in Chemistry 2008". 2008-10-08. http://nobelprize.org/nobel_prizes/chemistry/laureates/2008/. Retrieved 2008-10-08.
- ^ Shaner N, Steinbach P, Tsien R (2005). "A guide to choosing fluorescent proteins" (PDF). Nat Methods 2 (12): 905–9. doi:10.1038/nmeth819. PMID 16299475. http://tsienlab.ucsd.edu/Publications/Shaner%202005%20Nature%20Methods%20-%20Choosing%20fluorescent%20proteins.pdf.
- ^ Heim R, Cubitt A, Tsien R (1995). "Improved green fluorescence" (PDF). Nature 373 (6516): 663–4. doi:10.1038/373663b0. PMID 7854443. http://tsienlab.ucsd.edu/Publications/Heim%201995%20Nature%20-%20Improved%20GFP.PDF.
- ^ Thastrup O, Tullin S, Kongsbak Poulsen L, Bjørn S (2001). "Fluorescent Proteins". US patent. http://patft.uspto.gov/netacgi/nph-Parser?Sect2=PTO1&Sect2=HITOFF&p=1&u=%2Fnetahtml%2FPTO%2Fsearch-bool.html&r=1&f=G&l=50&d=PALL&RefSrch=yes&Query=PN%2F6172188.
- ^ Shelley R. McRae, Christopher L. Brown and Gillian R. Bushell (May 2005). "Rapid purification of EGFP, EYFP, and ECFP with high yield and purity". Protein Expression and Purification 41 (1): 121–127. doi:10.1016/j.pep.2004.12.030. PMID 15802229.
- ^ Pédelacq J, Cabantous S, Tran T, Terwilliger T, Waldo G (2006). "Engineering and characterization of a superfolder green fluorescent protein". Nat Biotechnol 24 (1): 79–88. doi:10.1038/nbt1172. PMID 16369541.
- ^ Miesenböck G, De Angelis D, Rothman J (1998). "Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins". Nature 394 (6689): 192–5. doi:10.1038/28190. PMID 9671304.
- ^ Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ (2004). "Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators". J Biol Chem 279 (13): 13044–53. doi:10.1074/jbc.M312846200. PMID 14722062.
- ^ Zacharias DA, Violin JD, Newton AC, Tsien RY (2002). "Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells". Science 296 (5569): 913–16. doi:10.1126/science.1068539. PMID 11988576.
- ^ Ormö M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ (September 1996). "Crystal structure of the Aequorea victoria green fluorescent protein". Science 273 (5280): 1392–5. doi:10.1126/science.273.5280.1392. PMID 8703075.
- ^ Yuste R (2005). "Fluorescence microscopy today". Nat Methods 2 (12): 902–4. doi:10.1038/nmeth1205-902. PMID 16299474.
- ^ Gunkel M, Erdel F, Rippe K, Lemmer P, Kaufmann R, Hörmann C, Amberger R, Cremer C (June 2009). "Dual color localization microscopy of cellular nanostructures". Biotechnol J 4 (6): 927–38. doi:10.1002/biot.200900005. PMID 19548231.
- ^ Chudakov D, Lukyanov S, Lukyanov K (2005). "Fluorescent proteins as a toolkit for in vivo imaging". Trends Biotechnol 23 (12): 605–13. doi:10.1016/j.tibtech.2005.10.005. PMID 16269193.
- ^ Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW (November 2007). "Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system". Nature 450 (7166): 56–62. doi:10.1038/nature06293. PMID 17972876.
- ^ Baker BJ, Mutoh H, Dimitrov D, Akemann W, Perron A, Iwamoto Y, Jin L, Cohen LB, Isacoff EY, Pieribone VA, Hughes T, Knöpfel T (August 2008). "Genetically encoded fluorescent sensors of membrane potential". Brain Cell Biol 36 (1–4): 53–67. doi:10.1007/s11068-008-9026-7. PMC 2775812. PMID 18679801. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2775812.
- ^ Adesnik H, Nicoll RA, England PM (December 2005). "Photoinactivation of native AMPA receptors reveals their real-time trafficking". Neuron 48 (6): 977–85. doi:10.1016/j.neuron.2005.11.030. PMID 16364901.
- ^ Lakadamyali M, Rust MJ, Babcock HP, Zhuang X (August 2003). "Visualizing infection of individual influenza viruses". Proc. Natl. Acad. Sci. U.S.A. 100 (16): 9280–5. doi:10.1073/pnas.0832269100. PMC 170909. PMID 12883000. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=170909.
- ^ Joo KI, Wang P (October 2008). "Visualization of Targeted Transduction by Engineered Lentiviral Vectors". Gene Ther. 15 (20): 1384–96. doi:10.1038/gt.2008.87. PMC 2575058. PMID 18480844. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2575058.
- ^ Remy S, Tesson L, Usal C, Menoret S, Bonnamain V, Nerriere-Daguin V, Rossignol J, Boyer C, Nguyen TH, Naveilhan P, Lescaudron L, Anegon I (January 2010). "New lines of GFP transgenic rats relevant for regenerative medicine and gene therapy". Transgenic Res 19 (5): 745–63. doi:10.1007/s11248-009-9352-2. PMID 20094912.
- ^ Stepanenko OV, Verkhusha VV, Kuznetsova IM, Uversky VN, Turoverov KK (August 2008). "Fluorescent Proteins as Biomarkers and Biosensors: Throwing Color Lights on Molecular and Cellular Processes". Curr. Protein Pept. Sci. 9 (4): 338–69. doi:10.2174/138920308785132668. PMC 2904242. PMID 18691124. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2904242.
- ^ Elliott G, McGrath J, Crockett-Torabi E (2000). "Green fluorescent protein: A novel viability assay for cryobiological applications". Cryobiology 40 (4): 360–369. doi:10.1006/cryo.2000.2258. PMID 10924267.
- ^ Gather, Malte C.; Yun, Seok Hyun (2011). "Single-cell biological lasers". Nature Photonics 5 (7): 406. doi:10.1038/nphoton.2011.99. http://www.nature.com/nphoton/journal/vaop/ncurrent/full/nphoton.2011.99.html. Retrieved 2011-06-13.
- ^ Matson, John (2011). "Green Fluorescent Protein Makes for Living Lasers". Scientific American. http://www.scientificamerican.com/article.cfm?id=biological-laser-cell. Retrieved 2011-06-13.
- ^ Voss-Andreae, J (2005). "Protein Sculptures: Life's Building Blocks Inspire Art". Leonardo 38: 41–45. doi:10.1162/leon.2005.38.1.41.
- ^ Pawlak, Alexander (2005). "Inspirierende Proteine". Physik Journal 4: 12.
- ^ "Julian Voss-Andreae Sculpture". http://www.julianvossandreae.com/. Retrieved 2007-06-14.
- ^ Eduardo Kac. "GFP Bunny". http://www.ekac.org/gfpbunny.html#gfpbunnyanchor.
- ^ [1] Glow-In-The Dark NeonMice
- ^ Scientists in Taiwan breed fluorescent green pigs
- ^ Wongsrikeao P, Saenz D, Rinkoski T, Otoi T, Poeschla E (2011). "Antiviral restriction factor transgenesis in the domestic cat". Nature Methods 8 (10): 853–9. doi:10.1038/nmeth.1703. PMID 21909101. http://www.nature.com/nmeth/journal/vaop/ncurrent/full/nmeth.1703.html.
Further reading
- Pieribone V, Gruber D (2006). Aglow in the Dark: The Revolutionary Science of Biofluorescence. Cambridge: Belknap Press. ISBN 0674019210. OCLC 60321612. Popular science book describing history and discovery of GFP
- Zimmer M (2005). Glowing Genes: A Revolution In Biotechnology. Buffalo, NY: Prometheus Books. ISBN 1591022533. OCLC 56614624.
External links
- Video: Roger Tsien Gives Nobel Lecture at UC San Diego
- A comprehensive article on fluorescent proteins at Scholarpedia
- Introduction to fluorescent proteins
- History, uses, and structure of GFP
- Brief summary of landmark GFP papers
- Interactive Java applet demonstrating the chemistry behind the formation of the GFP chromophore
- Tsien Lab @ UCSD
- Video of 2008 Nobel Prize lecture of Roger Tsien on fluorescent proteins
- Excitation and emission spectra for various fluorescent proteins
- Timeline
- Some factoids about Aequorea, the jellyfish source of GFP
- Video introduction to GFP, from the "Secrets of the Sequence" educational video series
- Green Fluorescent Protein Chem Soc Rev themed issue dedicated to the 2008 Nobel Prize winners in Chemistry, Professors Osamu Shimomura, Martin Chalfie and Roger Y. Tsien
Proteins: key methods of study Experimental Protein purification · Green fluorescent protein · Western blot · Protein immunostaining · Protein sequencing · Gel electrophoresis/Protein electrophoresis · Protein immunoprecipitation · Peptide mass fingerprinting · Dual polarization interferometry · Microscale thermophoresis · Chromatin immunoprecipitationBioinformatics Protein structure prediction · Protein–protein docking · Protein structural alignment · Protein ontology · Protein–protein interaction predictionAssay Display techniques Super resolution microscopy Vertico SMICategories:- Protein methods
- Recombinant proteins
- Cell imaging
- Fluorescent dyes
Wikimedia Foundation. 2010.